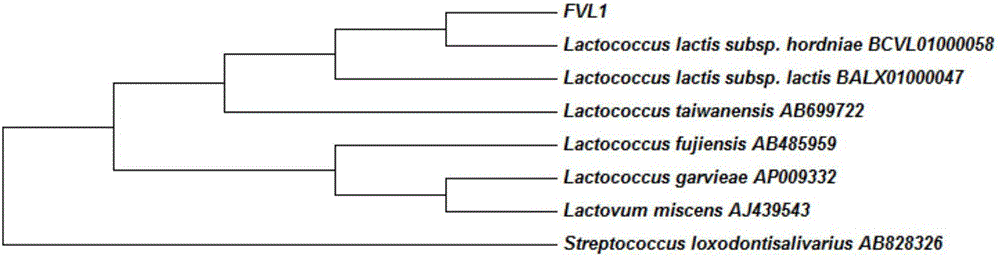
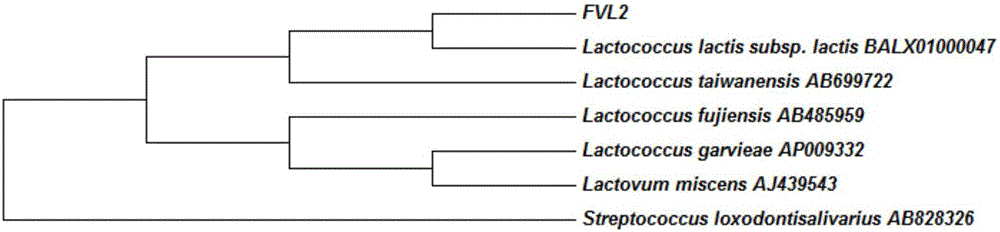
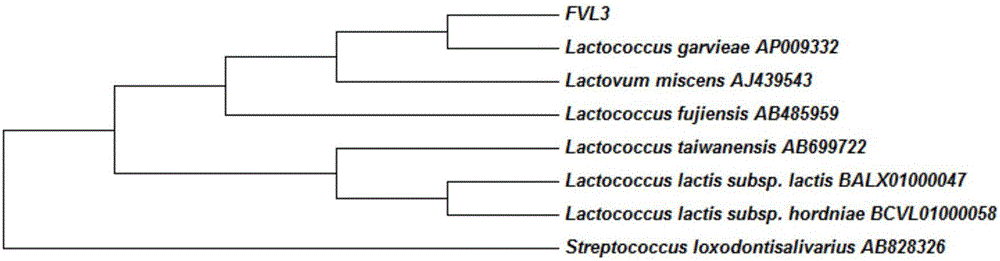
LAMP primer for detecting enoki mushroom surface lactococcus and detection method
A technology for Lactococcus and Flammulina velutipes, applied in biochemical equipment and methods, recombinant DNA technology, microbial measurement/inspection, etc., can solve the problems of not finding and detecting Lactococcus on the surface of Flammulina velutipes, achieve fast detection speed, easy observation, and method simple effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0056] Example 1 Bacteria Isolation and Identification Test on the Surface of Fresh Flammulina velutipes
[0057] (1) Test materials
[0058] Flammulina velutipes sample source: Fresh Flammulina velutipes were purchased from the supermarket Fa Supermarket at the South Gate of the Chinese Academy of Agricultural Sciences, packed in plastic wrap on trays.
[0059] (2) Preparation of beef extract-peptone medium: the composition and proportion of the medium are: 3g beef extract, 10g peptone, 5g NaCl, 20g agar powder, 20g glucose, 1000ml distilled water, pH 7.0-7.2, sterilized at 121°C 20min.
[0060] (3) Test method
[0061] (1) Sampling: Aseptically cut the surface tissue of Flammulina velutipes into pieces, mix them evenly, take 10g and put them into a sterile bag.
[0062] (2) Preparation of bacterial suspension: add 90ml of sterile saline to the sterile bag, and vibrate for 30s with a pulse homogenizer (Guangdong Huankai Microbial Technology Co., Ltd.: ShockMixer-1) to obta...
Embodiment 2
[0084] Example 2 Screening test of primers for three dominant Lactococcus strains on the surface of Flammulina velutipes
[0085] (1) Strain material
[0086] Among the 22 bacterial strains (see Table 1) isolated from the surface of Flammulina velutipes in Example 1, Nos. 1 to 3 are bacteria of the genus Lactococcus, and other bacterial strains are used as specificity test controls.
[0087] (2) Test method
[0088] (1) Determine candidate genes: take Lactococcus lactis subsp. The target gene, wherein the gene sequence of rpoA is shown in SEQ ID No.14.
[0089] (2) Design primers: according to the LAMP primer design software (URL: http: / / primerexplorer.jp / e / ), the DNA to be amplified is divided into six independent regions, and the primers required for the LAMP reaction (inner primers FIP and BIP, outer primers F3 and B3, loop primers LB and LF) are designed according to these six regions, each Five sets of LAMP primers were designed for each target gene, and a total of ...
Embodiment 3
[0106] Example 3 rpo289 primer set sensitivity detection test
[0107] (1) Test materials
[0108] Bacterial strain: Lactococcus lactis subsp. lactis (Lactococcus lactis subsp. lactis) FVL2 strain isolated from the surface of Flammulina velutipes.
[0109] (2) Reagent preparation:
[0110] The kit for LAMP nucleic acid amplification was purchased from Beijing Lanpu Biotechnology Co., Ltd. The kit mainly includes: 20mM Tris-HCl (pH 8.8), 10mM KCl, 10mM (NH 4 ) 2 SO 4 , 0.1% Tween20, 0.8M Betaine, 8mM MgSO 4 , 1.4mM dNTPs and 8U Bst DNA polymerase. Fluorescent visual detection reagents for detecting LAMP reactions were purchased from Beijing Lanpu Biotechnology Co., Ltd. Primers were synthesized by Sangon Biotech Co., Ltd. Takara ExTaq Mix was purchased from Bao Biological Engineering Co., Ltd. TIANGEN bacterial genome extraction kit.
[0111] (3) Test method
[0112] (1) Genome concentration sensitivity experiment
[0113] (a) Template preparation: Lactococcus lactis...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com