A real-time fluorescent PCR method of mgb probe for detecting hla-a*31:01 allele and its combination of primers and probes
A technology of HLA-A and alleles, applied in the field of alleles, can solve the problems of unsatisfactory detection requirements and flexibility, secondary pollution, false positives, etc., to save experimental time and consumables, reliable results, and high specificity Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Example 1 Detection of HLA-A*31:01 alleles by MGB probe method
[0047] 1. DNA sample extraction and dilution
[0048] After collecting venous blood in vacuum blood collection tubes anticoagulated with ethylenediaminetetraacetic acid (EDTA) according to conventional methods, DNA was extracted using the QIAamp DNA Mini Blood Kit (Qiagen, Germany) kit; the extracted DNA was concentrated using NanoDrop 2000 Determination (A 260 / 280 =1.95~2.15). Using the above method, extract 100 cases of Lantian Han DNA samples and measure the relative concentration, and then use PCR grade H 2 O Dilute the sample to 10 ng / μL.
[0049] 2. Design primers and probes
[0050] In the area where polymorphic sites are concentrated, specific primers for HLA-A*31:01 were designed by ARMS method, upstream primer Fp: 5'-GAGCCAGAGGATGGAGCC-3', downstream primer Rp: 5'-CCAGGTCCACTCGGTCtA-3', And the matching fluorescent probe probe1:5'-FAM-aGGCCtGAGTATTGGGAC–MGB-3', probe2:5'-VIC-CCTTCACaTTCCGTGT...
Embodiment 2
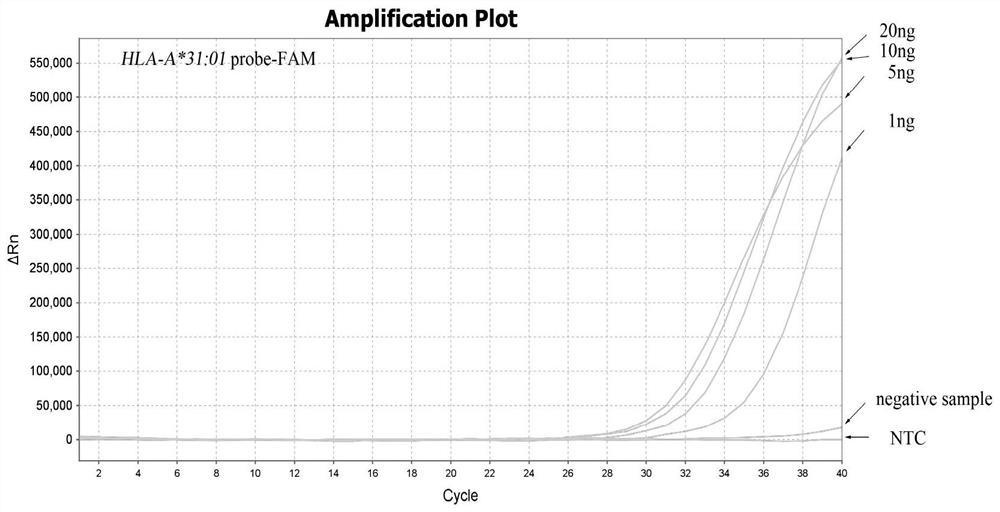
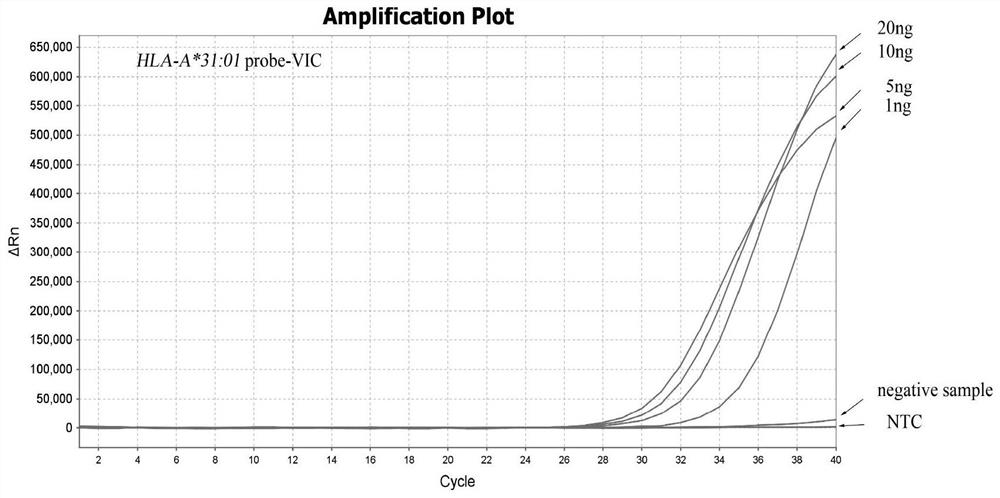
[0057] Example 2 Sensitivity detection of HLA-A*31:01 allele typing
[0058] 1. Dilution of DNA samples
[0059] Take HLA-A*31:01 standard DNA as the test sample, the concentration is 2ng / μL; 2 O Serially diluted samples, namely 1:2, 1:4, 1:20, 1:40, 1:80; the DNA concentrations were: 10ng / μL, 5ng / μL, 1ng / μL, 0.5ng / μL, 0.25 ng / μL.
[0060] 2. Design primers and probes
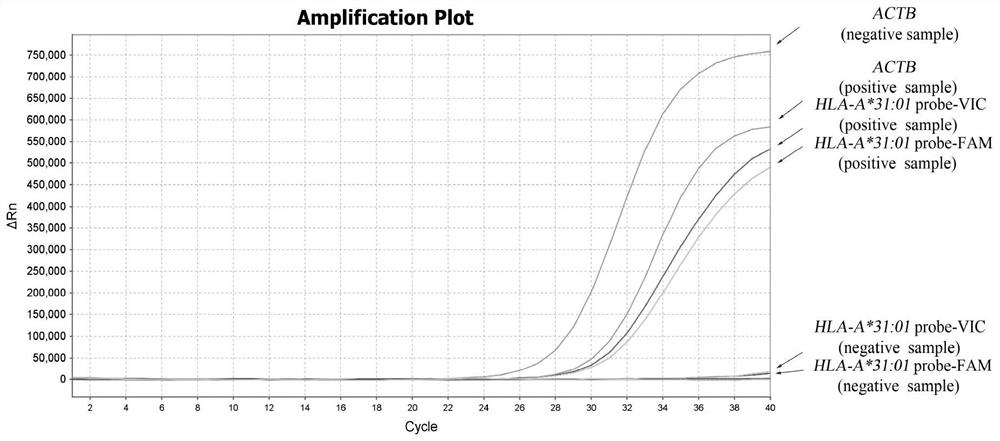
[0061]Design specific primers for HLA-A*31:01 by ARMS method, upstream primer Fp:5'-GAGCCAGAGGATGGAGCC-3', downstream primer Rp:5'-CCAGGTCCACTCGGTCtA-3', and matching fluorescent probe probe1:5' -FAM-aGGCCtGAGTATTGGGAC-MGB-3', probe2:5'-VIC-CCTTCACaTTCCGTGTCTC-MGB-3'; In addition, internal reference primers were designed on the ACTB gene, upstream primer Actin-F: 5'-CAGCAGATGTGGATCAGCAAG-3', downstream primer Actin -R: 5'-GCATTTGCGGTGGACGAT-3', and the matching fluorescent probe Actin-probe: 5'-CY5-AGGAGTATGACGAGTCCGGCCCC-BHQ2-3'.
[0062] Among them, FAM is 6-carboxyfluorescein; VIC is 4,7,2′-trichloro-7′...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com