Method for visually marking genome sites
A genome and labeling technology, applied in the biological field, can solve problems such as the inability to accurately label genomic elements and special sites in human cells, low signal-to-noise ratio, and genome damage.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
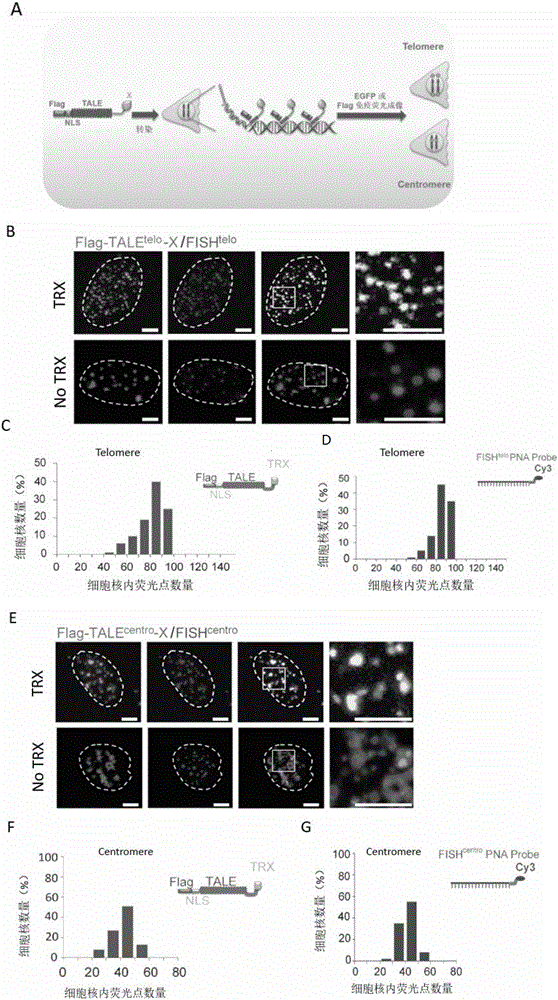
[0098] Example 1, Establishment of Genome Visualization Labeling Technology (TTALE) for Accurately Marking Telomere and Centromere in Cell Genome
[0099] 1. Construction of the expression vector TTALE for fusion expression of TALE and TRX
[0100] 1. TALE vectors that recognize telomeres and centromeres
[0101] Refer to the literature "Zhang, F., et al., Efficient construction of sequence-specific TAL effector for modulating mammalian transcription. Nat Biotechnology, 2011.29(2): p.149-53, using the method in Golden Gate Assembly" using TALE Toolbox Kit (Addgene, USA, Cat. No. 1000000019) to construct TALE vectors recognizing telomeres and TALE vectors recognizing centromeres, respectively.
[0102] (1) TALE carrier that recognizes telomeres
[0103] The nucleotide sequence of the TALE vector recognizing telomeres is shown in Sequence 1. Telomere-recognizing TALE vector expresses fusion protein TALE telo , the fusion protein TALE tel The target sequence of the identifie...
Embodiment 2
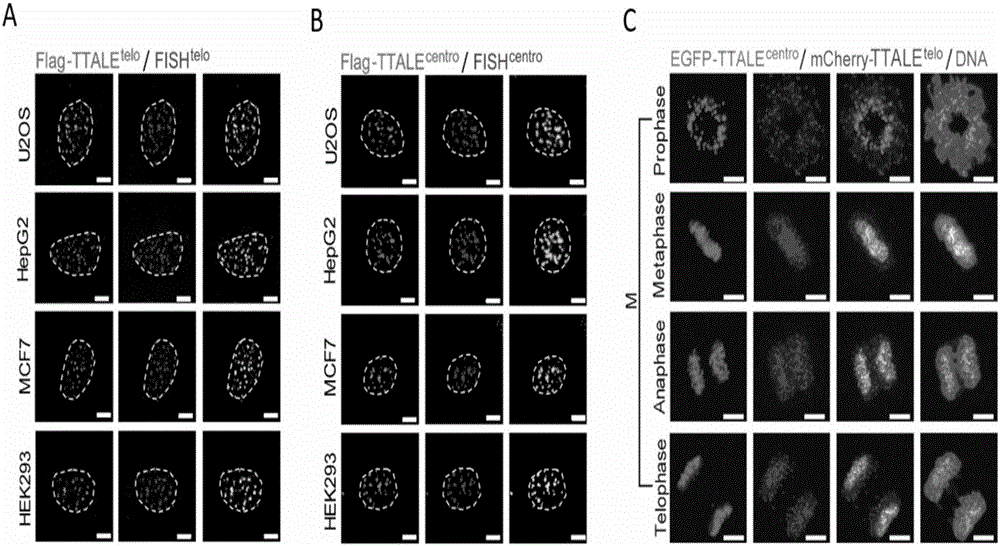
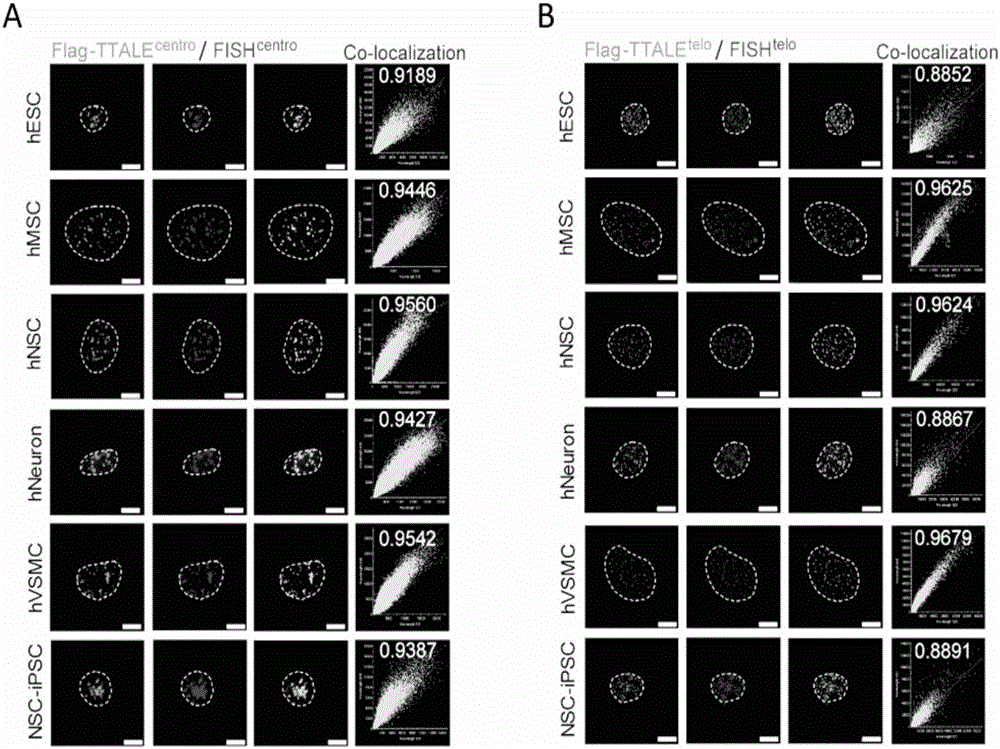
[0119] Example 2, Application of TTALE in Accurately Marking Telomere, Centromere, Nucleolus Organization Region Ribosomal RNA Coding Sequence (NOR-rDNA) and Coding Gene Locus (MUC4) in Different Types of Human Cells
[0120] 1. Application of TTALE in precise labeling of telomeres and centromeres in different types of human cells
[0121] The U2OS cells in Example 1 were replaced with the following cells respectively: human tumor cell lines (MCF7 and HepG2, American ATCC Company, product numbers HTB-22 and HB-8065 respectively), human embryonic kidney cell lines (HEK293, American ATCC Company , product number is CRL-1573), human pluripotent stem cells (embryonic stem cells hESC, Wicell Company, product number WA-09), human pluripotent stem cells (induced pluripotent stem cells iPSC, Wicell Company, product number IISH6i-CML17), adult stem cells (Mesenchymal stem cells, Lonza company, product number is PT-2501), adult stem cells (neural stem cells hNSC, Wicell company, product...
Embodiment 3
[0151] Example 3. Application of TTALE to mark telomeres in different cell aging models in visualizing the dynamic changes of cell aging process
[0152] 1. Preparation of TTALE vector for telomere recognition of EGFP fusion expression
[0153] The EGFP coding gene sequence shown in the 7382-8098 position of sequence 13 is replaced with the TTALE carrier (mCherry-TTALE telo ) in the mCherry coding gene sequence to obtain the TTALE carrier (EGFP-TTALE telo ), the N-terminus of the fusion protein has a FLAG tag sequence and a nuclear localization sequence NLS.
[0154] 2. Human mesenchymal stem cells of different aging models labeled with TTALE carrier expressing EGFP fusion and recognizing telomeres
[0155] (1) Establishment of human mesenchymal stem cells in different aging models
[0156] 1) Preparation of wild-type human mesenchymal stem cells (WT-MSC) and WRN gene-deleted human mesenchymal stem cells (WS-MSC)
[0157] In the present invention, wild-type human embryonic...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com