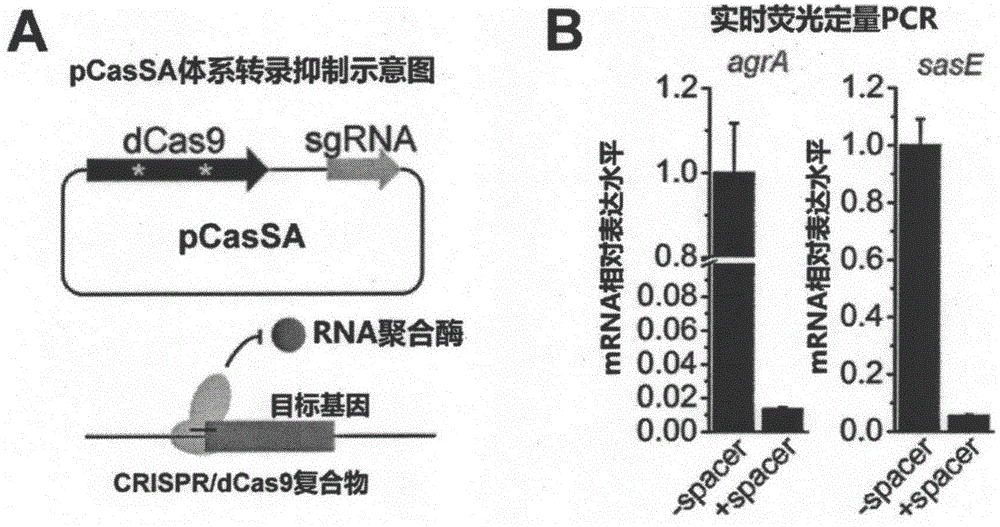
pCasSA plasmid and application thereof
A plasmid and golden yellow technology, applied in the field of pCasSA plasmid, can solve the problem of genome editing without Staphylococcus aureus, achieve high-efficiency transcription inhibition and broad application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Embodiment 1: pCasSA plasmid construction:
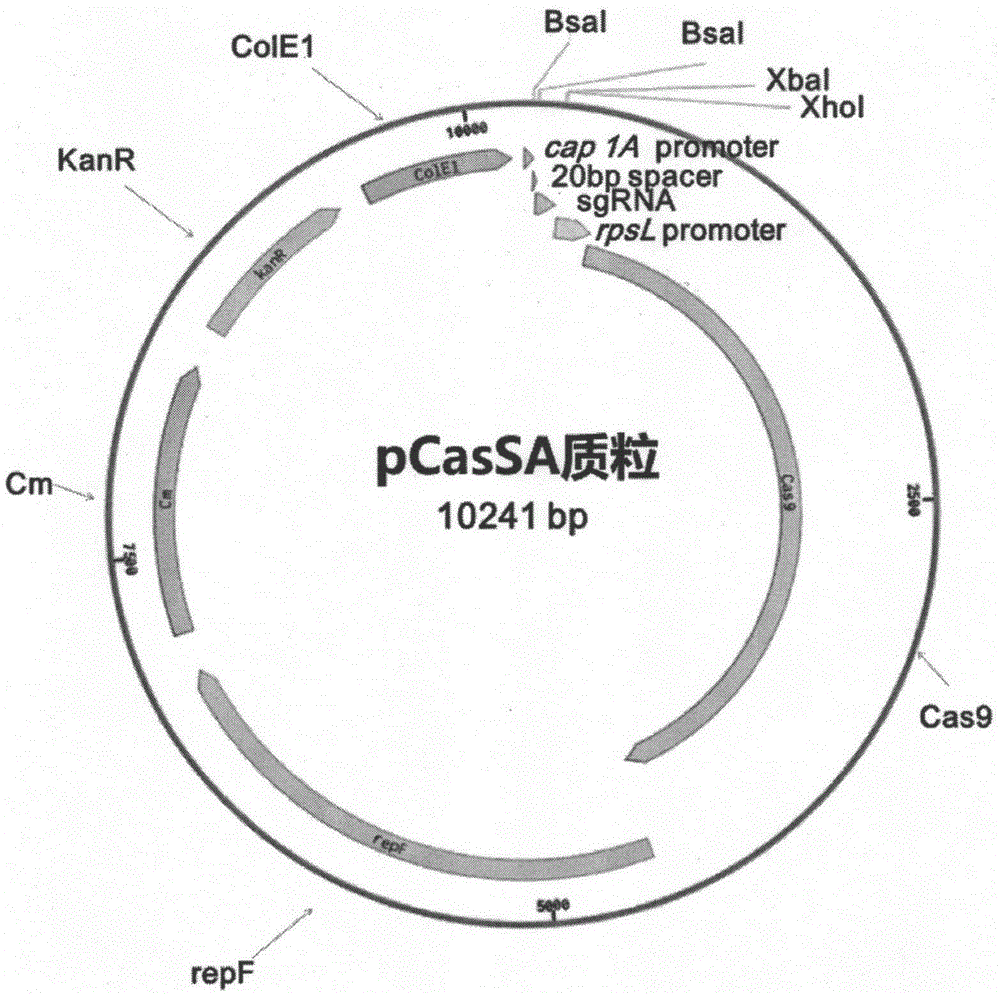
[0033] The composition of the pCasSA plasmid is attached figure 1Shown, its sequence is SEQ ID NO:1. The specific construction method of the pCasSA plasmid is as follows:
[0034] (1) The genome of Staphylococcus aureus Newman strain was extracted using the Ezup Column Bacterial Genomic DNA Extraction Kit produced by Sangon Bioengineering (Shanghai) Co., Ltd., and the specific steps were carried out according to the operation manual of the kit. The promoter of the rpsL gene was amplified by PCR using the genome of the Newman strain as a template; the Cas9 protein gene was amplified by PCR from the pCas9 plasmid; the temperature-sensitive repF replicon and the chloramphenicol-resistant gene were amplified by PCR respectively from the pKOR1 plasmid. Sex gene fragment; DNA fragment containing ColE1 replicon and kanamycin resistance gene was obtained by PCR amplification from pCRISPR plasmid.
[0035] The primer sequences used...
Embodiment 2
[0068] Example 2: pCasSA achieves efficient gene knockout in various strains of Staphylococcus aureus:
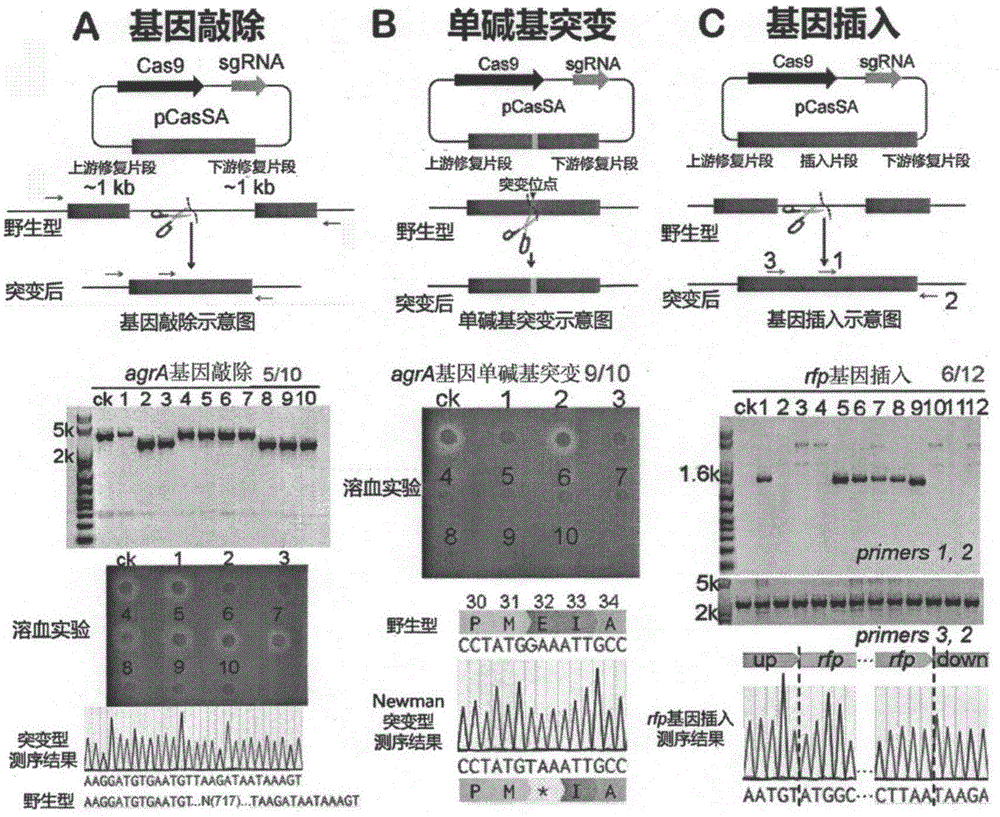
[0069] The pCasSA plasmid can be used to efficiently knock out different genes in various strains of Staphylococcus aureus. In the experiment, the agrA gene was selected as an example, and gene knockout experiments were carried out in Staphylococcus aureus RN4220, Newman and USA300 strains. attached figure 2 A is a schematic diagram of gene knockout in Staphylococcus aureus using pCasSA system, and PCR verification, hemolysis experiment and DNA sequencing results of agrA gene knockout in Newman strain.
[0070](1) First select a DNA fragment of the first 20 bases of a certain NGG (N is any base) sequence on the target gene (these 20 bases are called spacers, NGG is not included in it, and the GC ratio of the spacer Preferably at 40%-60%). The feature of this step is that in order to enable the spacer fragment to be inserted into the pCasSA plasmid, GAAA needs to be adde...
Embodiment 3
[0108] Example 3: pCasSA achieves efficient single-base mutations in various strains of Staphylococcus aureus:
[0109] The pCasSA plasmid can be used to perform highly efficient single-base mutations in different genes in various strains of Staphylococcus aureus. In the experiment, the agrA gene was selected as an example, and the gene single base mutation experiment was carried out in Staphylococcus aureus RN4220 and Newman strains. attached figure 2 B is a schematic diagram of using the pCasSA system to carry out single-base mutation of the gene in Staphylococcus aureus, and the hemolysis experiment and DNA sequencing results of the single-base mutation of the agrA gene in the Newman strain.
[0110] (1) Select the spacer of the target gene, and design primers to construct the plasmid. In the experiment, the guanine G at position 94 in the agrA gene was selected and mutated into thymine T. After the mutation, the TGG on the original agrA gene becomes TGT, and the Cas9 p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com