Escherichia coli gene engineering bacterium and method for producing L-threonine by using escherichia coli gene engineering bacterium
A technology of genetically engineered bacteria and Escherichia coli, applied in the field of genetic engineering, can solve problems such as affecting the synthesis and accumulation of L-threonine, affecting the normal metabolism and growth of cells, etc., to improve the conversion rate of sugar and acid, and enhance the yield and sugar acid. Conversion rate, ease of use effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
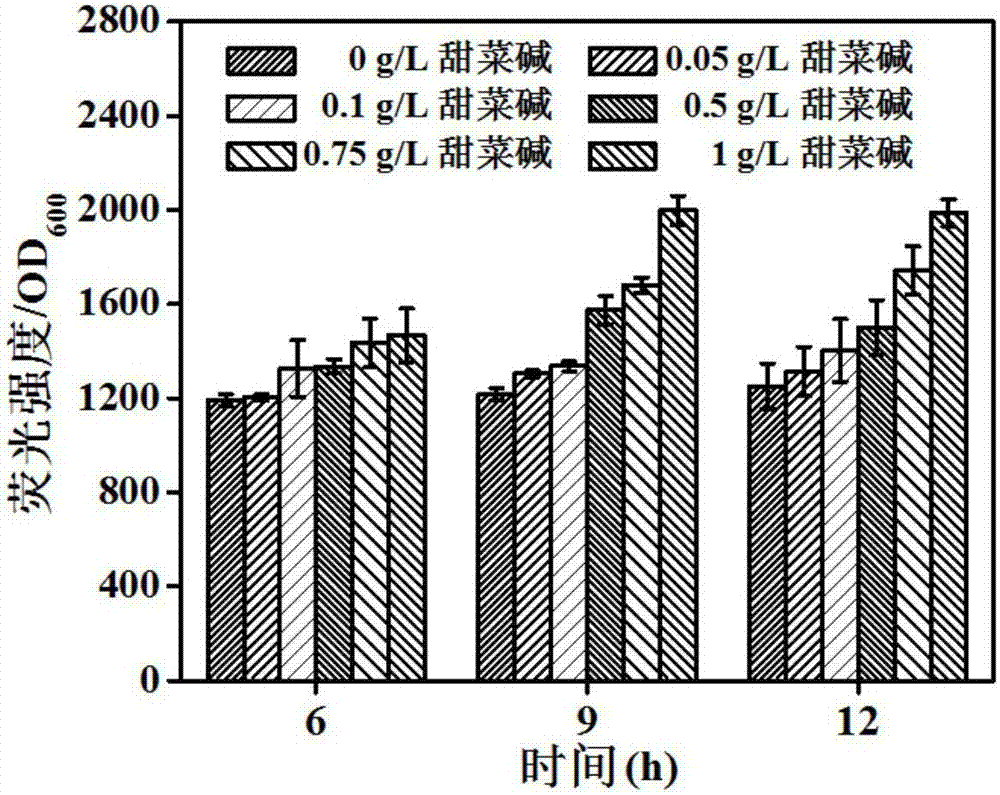
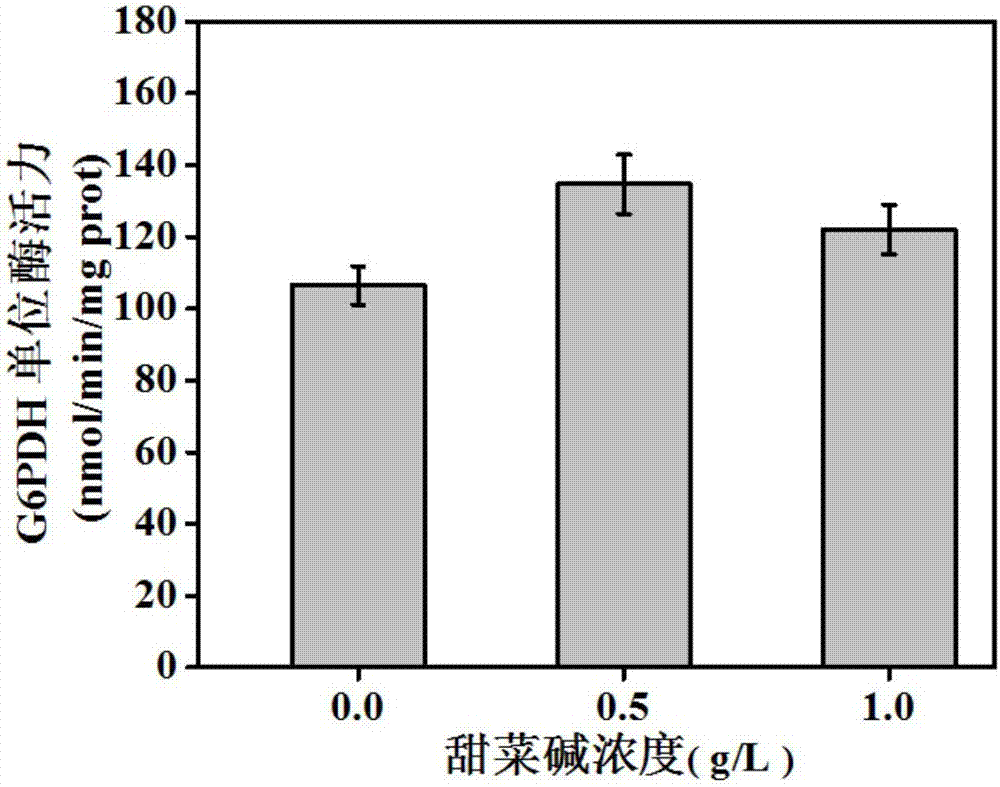
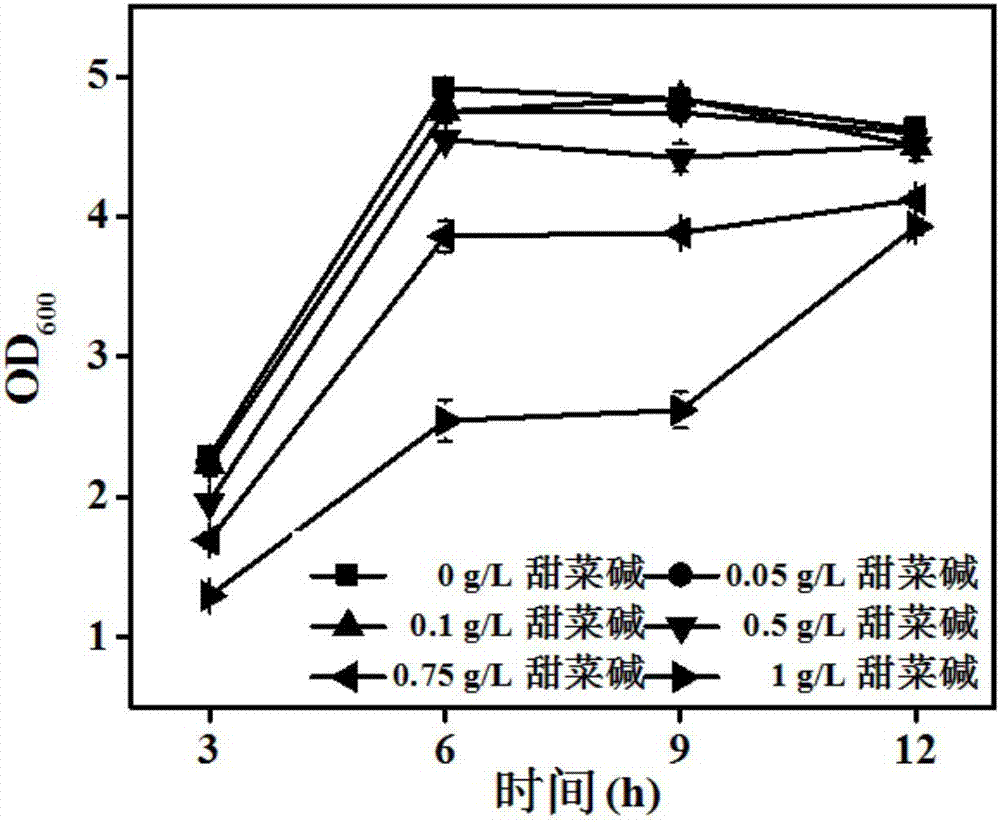
[0052] Example 1: Betaine enhances P zwf expression strength test
[0053] 1. Construction of a recombinant vector comprising the zwf gene promoter and the green fluorescent protein expression gene gfp
[0054] (1) Transformation of vector pUC19
[0055] Transform the vector pUC19 by designing reverse amplification primers pUC19-F / pUC19-R to remove the entire Lac operon (see Figure 4 ), the transformed vector fragment was named pUC19-L (SEQ ID No.1);
[0056] (2) Amplification of zwf gene promoter and gfp gene and construction of overlapping fragments
[0057] Using the zwf promoter sequence (SEQ ID No.2) in the Escherichia coli MG1655 genome as a template to design the amplification primer P zwf –up and P zwf -down; use the gfp (SEQ ID No.3) gene sequence on the pEGFP-N1 plasmid as a template to design amplification primers gfp–up and gfp–down, primer P zwf –down and gfp–up contain a 20bp overlapping sequence, primer P zwf The 5' ends of –up and gfp-down each contain ...
Embodiment 2
[0089] Example 2: P of E. coli THRD ppc replace with P zwf
[0090] 1. ppc gene promoter P ppc Upper and lower homology arms, Cm gene and zwf gene promoter P zwf Amplification of
[0091] The promoter P of the ppc gene (SEQ ID No.20) on the genome of Escherichia coli MG1655 ppc The upper and lower homology arm sequences are used as templates to design primers ppc-up-1 / 2 and ppc-down-1 / 2;
[0092]Primers zwf-1 and zwf-2 were designed using the zwf promoter sequence on the genome of Escherichia coli MG1655 as a template;
[0093] Cm fragment primers Cm-up and Cm-down were designed using plasmid pKD3 as a template.
[0094] The high-fidelity enzyme PrimeSTAR HS DNA Polymerase was used to amplify the upper and lower homology arm sequences of the ppc gene promoter and the zwf promoter sequence respectively, and the target band was obtained after electrophoresis verification (see Figure 5 ), using the OMEGA PCR product purification kit of Guangzhou Feiyang Bioengineering Co....
Embodiment 3
[0101] Embodiment 3: Shake flask fermentation
[0102] Test strain: the bacterial strain obtained in Example 2; Protobacterium THRD;
[0103] (1) The test strains were activated by slant culture, cultured at 37°C for 16h, transferred once (second-generation slope activation), and cultured at 37°C for 10h;
[0104] (2) Use an inoculation loop to draw the above-mentioned second-generation slant two-ring sludge into 30mL seed medium, and cultivate it to OD at 37°C and 200rpm 600 is 8;
[0105] (3) Inoculate the seed culture solution into the fermentation medium according to the inoculum size of 10%, cultivate at 37°C and 200rpm, start to add sugar after the lack of sugar, and betaine is added with sugar (80% glucose is added, containing 2.5 g / L betaine, add 1mL per 30mL fermentation system each time), and the fermentation ends for 28h;
[0106] Sugar deficiency standard: If there is no color change in the culture medium for more than 20 minutes (that is, the pH does not change...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com