Method for detecting gene nucleotide mutation site of rice ustilaginoidea virens cyp51
A technology of rice smut bacteria and mutation sites, applied in the field of molecular biology, can solve the problems of heavy workload, low sensitivity, and long time-consuming, and achieve the effect of heavy workload, high accuracy, and long cycle
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Example 1. Discovery of cyp51 gene and CYP51 protein mutation sites in rice smut.
[0034] 1. Strains: The resistant strains are F10-338C and F10-338D, and the sensitive strains are F10-338, JY13107, NH13018, SX13035, and NH13052.
[0035] 2. Method:
[0036] 1) Strain culture: culture the strain on PSA medium at 28°C for 5-7 days, take 7-10 bacterial cakes and shake them in PSB liquid medium for 5 days, filter the mycelium balls, and remove the cultured solution, freeze-dried in a freeze dryer and stored at -20°C.
[0037] 2) Genomic DNA of resistant strains and sensitive strains were extracted respectively.
[0038] 3) Genomic RNA of resistant strains and sensitive strains were extracted respectively.
[0039] 4) RNA reverse transcription.
[0040] 5) PCR amplification of lanosterol 14α-demethylase cyp51 gene
[0041] Table 1 Cloning primers of lanosterol 14α-demethylase cyp51 gene
[0042]
[0043] The primers used are S-4 / A-3 in Table 1.
[0044] PCR react...
Embodiment 2
[0058] Embodiment 2, the detection of mutation site in rice smut bacterium
[0059] Experiment 1. Primer design
[0060] Primers are listed in Table 2.
[0061] Table 2. Primers used in AS-PCR detection of resistance of rice smut to tebuconazole
[0062]
[0063] In this method, the last base of the downstream primer is consistent with the mutant (Table 2, primer Uv137AR), and the penultimate base introduces a mismatched base (Table 2, primers Uv137BR, Uv137CR, Uv137DR), and combined with different annealing Amplification at a higher temperature increases the specificity of the primers. The reaction conditions are as follows:
[0064] The PCR reaction system is as follows: 25 µL reaction system includes: 0.5 µL Taq DNA polymerase (2.5 U / µL), 2.5 µL 10×PCR buffer (containing Mg 2+ ), 2.0 μL 10mM dNTPs, 0.5 μL each primer, 1 μL template DNA, 18 μL ddH 2 O.
[0065] PCR reaction conditions: pre-denaturation at 94°C for 3min, 98°C for 10s, 44-56°C for 30s, 68°C for 45s, 3...
Embodiment 3
[0073] Embodiment 3, the rice false smut bacteria detection of resistance to tebuconazole
[0074] The PCR amplification system and reaction conditions were consistent with Experiment 1 in Example 2, and the annealing temperature was 51.6°C.
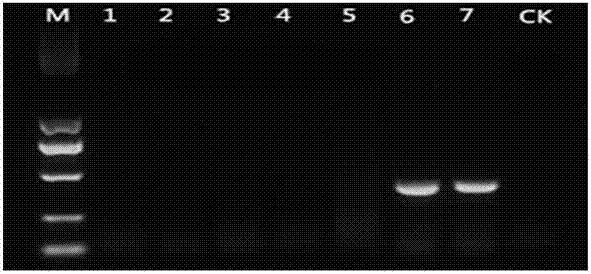
[0075] The rice curve balls to be tested are: rice curve balls produced by inoculating rice plants with resistant strains F10-338C and F10-338D respectively; rice curve balls produced by inoculating rice plants with sensitive strains F10-338, JY13107, NH13018 and SX13035 respectively. The rice curved balls were quickly ground with liquid nitrogen, and then the DNA molecules in the rice curved balls were extracted using a DNA extraction kit.
[0076] The result is as Figure 4 shown. The results showed that the primers Uv137F1 / Uv137DR could only amplify a specific 391 bp band (lane 1, 2) from the rice curve balls produced by inoculating rice plants with a strain resistant to tebuconazole, but not from a sensitive strain. Specific 391 b...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com