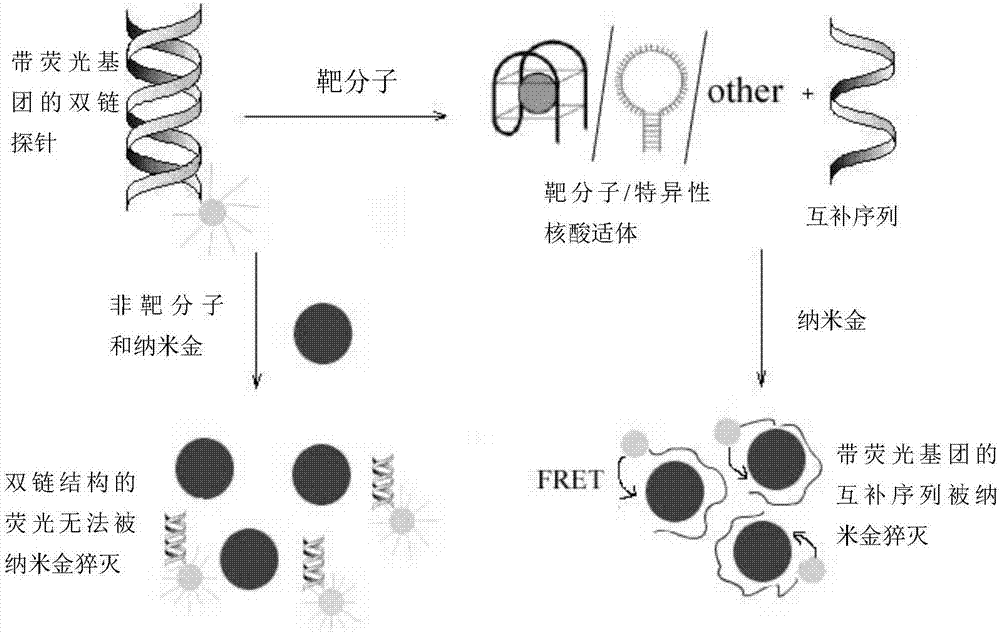
Method for detecting tumor marker
A tumor marker and nucleic acid aptamer technology, applied in the field of new nano-probes, can solve the problem of no significant decrease in fluorescence and achieve rapid detection results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Example 1 Nano gold-DNA interaction principle to the detection of prostate-specific antigen (PSA)
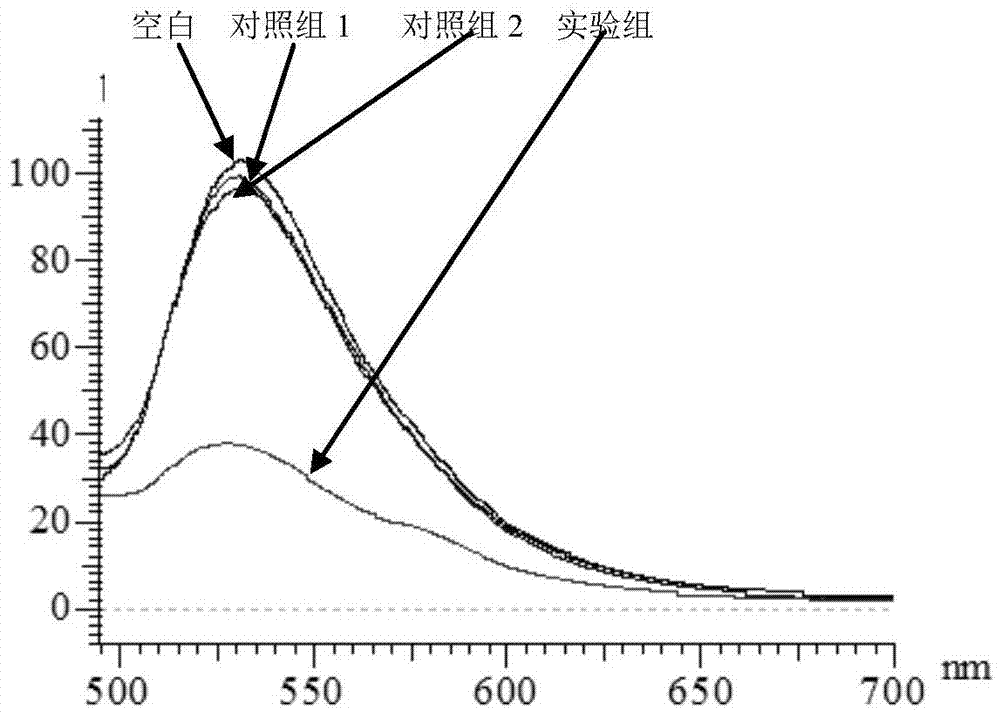
[0034]Steps: Take 2 μL of PSA-aptamer solution with a concentration of 100 μM and 2 μL of a cDNA solution with a concentration of 100 μM, add 18 μL of buffer solution (25 mM Tris, pH 8.2, 0.3 M NaCl) and fully hybridize to form a double-stranded probe (room temperature reaction for 30 minutes). Then, 2 μL of 1 mM PSA aqueous solution was added to the hybridization solution, and fully reacted at room temperature. At the same time, the group without adding PSA and adding 2 μL of water was used as blank, and the two groups of adding 2 μL of 1 mM AFP (N1) and CEA (N2) were used as control group 1 and control group 2, respectively. Then, 2 μL of the above reaction solution was added to 100 μL of 3.5 nM gold nano-solution, and a buffer solution (25 mM Tris, pH 8.2, 0.3 M NaCl) was used to make up the total volume of 0.2 mL, and the fluorescence spectra of the experimental grou...
Embodiment 2
[0036] Example 2 The detection of carcinoembryonic antigen (CEA) by the principle of nano-gold-DNA interaction
[0037] Steps: Take 2 μL of CEA-aptamer solution with a concentration of 100 μM and 2 μL of a cDNA solution with a concentration of 100 μM, add 18 μL of buffer solution (25 mM Tris, pH 8.2, 0.3 M NaCl) and perform sufficient hybridization to form a double-stranded probe (room temperature reaction for 30 minutes). Then, 2 μL of 0.1 mM CEA aqueous solution was added to the hybridization solution, and a sufficient reaction was carried out at room temperature. At the same time, a group without CEA and 2 μL of water was used as a blank, and a group with 2 μL of 1 mM PSA and AFP was used as a control. Then 2 μL of the above reaction solution was added to 10 μL of 3.5 nM gold nanometer solution, and the buffer solution was used to make up the total volume of 0.2 mL, and the fluorescence spectra of the experimental group and the control group were recorded.
[0038] Result...
Embodiment 3
[0039] Example 3 The detection of alpha-fetoprotein (AFP) by the principle of nano-gold-DNA interaction
[0040] Steps: Take 2 μL of AFP-aptamer solution with a concentration of 100 μM and 2 μL of cDNA solution with a concentration of 100 μM respectively, add 18 μL of buffer solution (20 mM Tris-acetic acid, pH 7.4, 140 mM NaCl, 1 mM CaCl 2 , 1 mM MgCl 2 ) to fully hybridize to form a double-stranded probe (30 minutes at room temperature). Then, 2 μL of an aqueous solution of AFP with a concentration of 0.05 mM was added to the hybridization solution, and a sufficient reaction was carried out at room temperature. At the same time, a group without adding AFP and adding 2 μL of water was used as a blank experiment. And a group to which 2 μL of 1 mM BSA was added was used as a control. Then 2 μL of the above reaction solution was added to 50 μL of 3.5 nM gold nanometer solution, and the buffer solution was used to make up the total volume of 0.2 mL, and the fluorescence spectr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com