A kind of enzyme immobilization method and its application
A technology for enzyme immobilization and immobilization of lipase, which is applied to the enzyme immobilization method and its application field, can solve the problems of low expression amount of exogenous protein, difficulty in separation and purification, etc., and achieves simple purification steps, cost saving, and improved thermal efficiency. Effects of stability and pH tolerance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
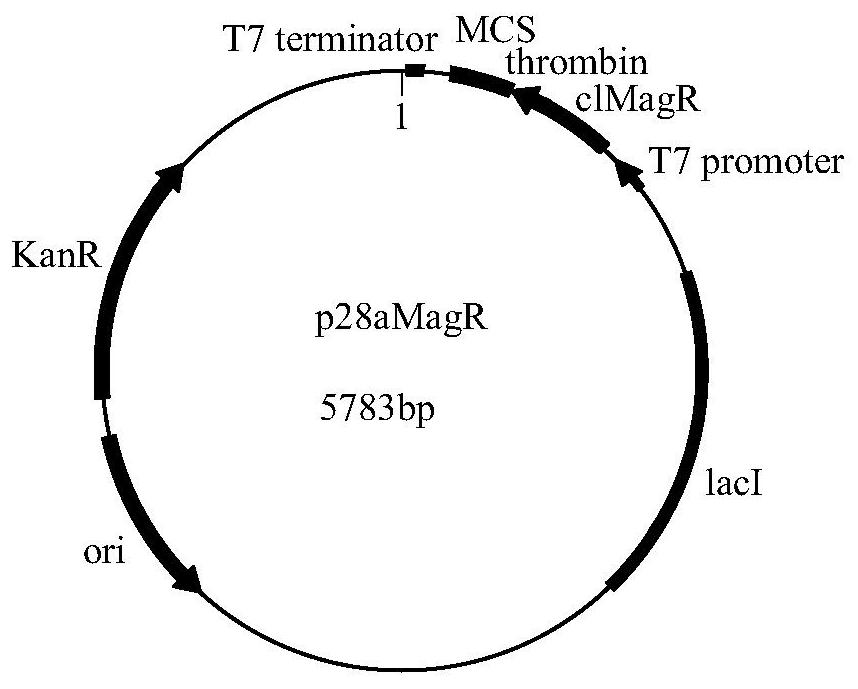
[0048] Example 1 Construction of pET28a-clMagR recombinant expression plasmid
[0049] 1) Synthesizing the nucleotide sequence of the clMagR gene shown in SEQ ID NO.1, which was synthesized by GenScript Biotechnology Co., Ltd.;
[0050] 2) Synthesize the following primers respectively:
[0051] Primer clMagRF: 5'CGC GGATCC ATGGCATCTAGCGCATCATC3', wherein the underlined part is the BamH I recognition site;
[0052] Primer clMagRR: 5'ACG GAGCTC GATGTTAAAGCTTTCTCCAC3', wherein the underlined part is the Sac I recognition site.
[0053] 3) Under the guidance of the above primers, perform PCR amplification using the sequence in step 1) as a template.
[0054] 4) The pET-28a (+) empty plasmid, and the gene with BamH I and SacI restriction sites obtained in step 3) were double-digested with BamH I and SacI respectively. Recover large fragments, connect them with T4 DNA ligase, transform Escherichia coli DH5a by heat shock method, screen with LB solid medium containing 50 μg / mL...
Embodiment 2
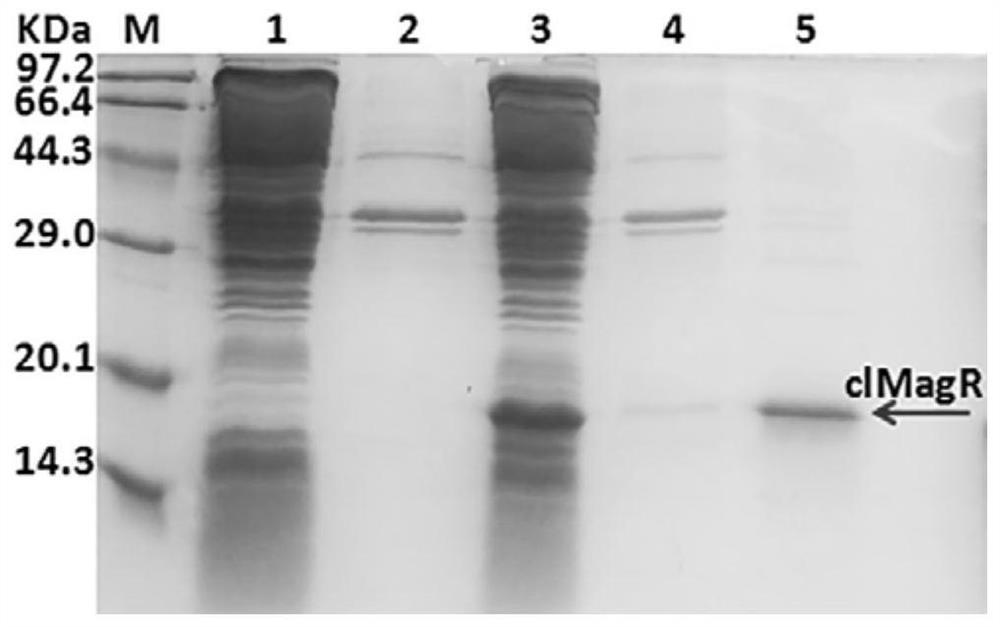
[0055] Example 2 Heterologous efficient soluble expression of magnetic protein and magnetic verification of magnetic protein
[0056] 1) The expression plasmid pET28a-clMagR obtained in Example 1 was transferred into Escherichia coli BL21(DE3) by heat shock method, and a single colony was picked and placed in 5 mL LB medium containing 50 μg / mL kanamycin for 37 Cultivate at ℃ for 8 hours, and then transfer to fresh 40ml LB medium at a ratio of 1% to continue to propagate. When OD600 reached 0.6-0.8, IPTG was added to a final concentration of 20 μM. After induction at 20°C for 13 hours, cells were collected and washed with pure water. The collected cells were resuspended in TBS buffer (20 mM Tris, 150 mM NaCl, pH 7.5) and sonicated for 15 minutes. The cell lysate was centrifuged, and the supernatant and precipitate were collected for SDS-PAGE electrophoresis.
[0057] 2) Take 0.5ml of the supernatant collected in step 1), add 0.1ml of Fe 3 o 4 -SiO 2 Nanoparticles (10mg ml...
Embodiment 3
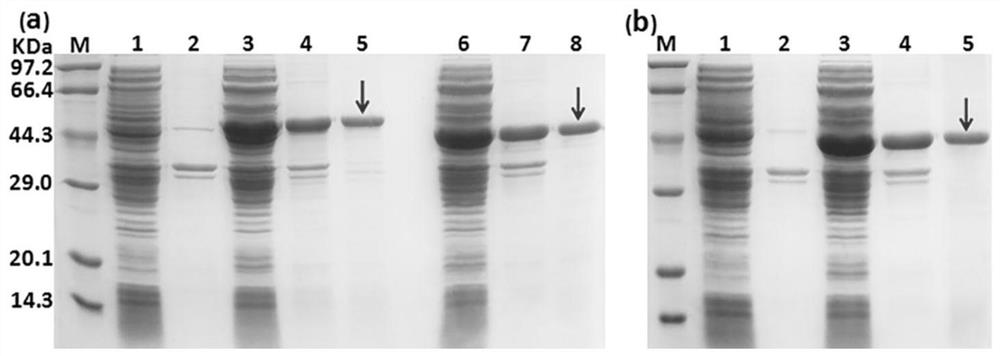
[0058] Example 3 linker design pattern
[0059] 1) Design and synthesize rigid linker and flexible linker, and primers linker1F, linker2F, linker1R / linker2R, GFP 1F / GFP1R, GFP2F / GFP2R, GFP3F / GFP3R;
[0060] Wherein, the gene sequence of the rigid linker is shown in SEQ ID NO.5, and the gene sequence of the flexible linker is shown in SEQ ID NO.6;
[0061] linker1F: 5'ATC GAGCTC AAAGCGAAACTGAAAGAGGA3', wherein the underlined part is the Sac I recognition site;
[0062] linker2F: 5'ATC GAGCTC GGCGGAGGTGGCTCTGGCGG3', wherein the underlined part is the Sac I recognition site;
[0063] linker1R / linker2R: 5'TTTCTCCTTTACTCATATGGCTGCCGCGCGG
[0064] CACCA3'
[0065] GFP1F / GFP2F: 5'CCATATGAGTAAAGGAGAAGAACT3'
[0066] GFP3F:5'ATC GAGCTC CTGGTGCCGCGCGGCAGCCATATGAGTAAAGGAGAAGAAC3', wherein the underlined part is the Sac I recognition site;
[0067] GFP1R / GFP2R / GFP3R: 5'AGT GCGGCCGC TTATTTGTATAGTTCATCCA3', wherein the underlined part is the Not I recognition site.
[0068] 2)...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com