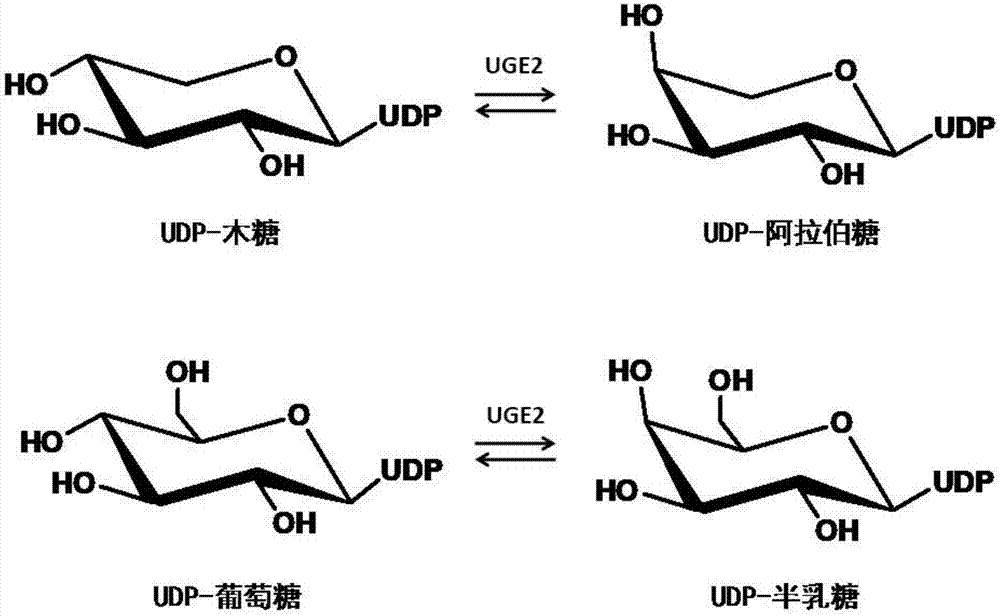
Uridine-5'-diphosphate galactose epimerase derived from ornithogalum caudatum, nucleotide sequence of uridine-5'-diphosphate galactose epimerase and application
A technology of galactose diphosphate and epimerase, applied in the direction of isomerase, application, biochemical equipment and methods, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Example 1. Transcriptome Sequencing and Sequence Analysis
[0030] After extracting the total RNA from the sterile bulb of Dieffenbachia tiger eye, using the mRNA as a template, the first cDNA strand was synthesized with six base random primers (randomhexamers), and then the second cDNA was synthesized by adding buffer, dNTPs, RNase H and DNA polymerase I After purification by QiaQuick PCR kit and elution with EB buffer, end repair, poly(A) was added and sequencing adapters were connected, then fragment size selection was performed by agarose gel electrophoresis, and finally PCR amplification was carried out to construct Good sequencing library with Illumina HiSeq TM 2000 for sequencing.
[0031]The original image data obtained by sequencing is converted into sequence data through base calling, that is, raw data or rawreads. Perform data filtering on raw reads, remove reads with adapters, duplicates, and low-quality sequencing, and obtain clean reads. Use the short ...
Embodiment 2
[0033] Example 2 OcUGE2 gene cloning
[0034] Take 100mg of aseptic bulbs of Dieffenbachia tiger's eye, freeze them quickly in liquid nitrogen, grind them into fine powder with a mortar, and use Trizol extraction to extract total RNA from the aseptic bulbs of Dieffenbachia tiger's eye. Using RT-PCR Kit (ReverTra-Plus-, TOYOBO company) to reverse transcribe the total RNA of Diofenbachia sterile bulbs into cDNA. The reverse transcription system and procedure are as follows: add 1 μg of total RNA to 20 μL system, RNase Free H 2 O 4 μL, Oligo(dT) 20 1 μL, after incubating at 65°C for 5 minutes, immediately place it on ice to cool, then add 4 μL of 5×RT buffer, 1 μL of RNase Inhibitor (10U / μL), 1 μL of dNTP Mixture (10mM) and ReverTra Ace to the above tube, at 30°C Incubate for 10 minutes, incubate at 42°C for 60 minutes, denature at 85°C for 5 minutes, and place on ice for 5 minutes to complete cDNA synthesis. The cDNA was stored at -20°C for later use.
[0035] Two pairs of ...
Embodiment 3
[0038] Example 3 Construction of OcUGE2 expression vector
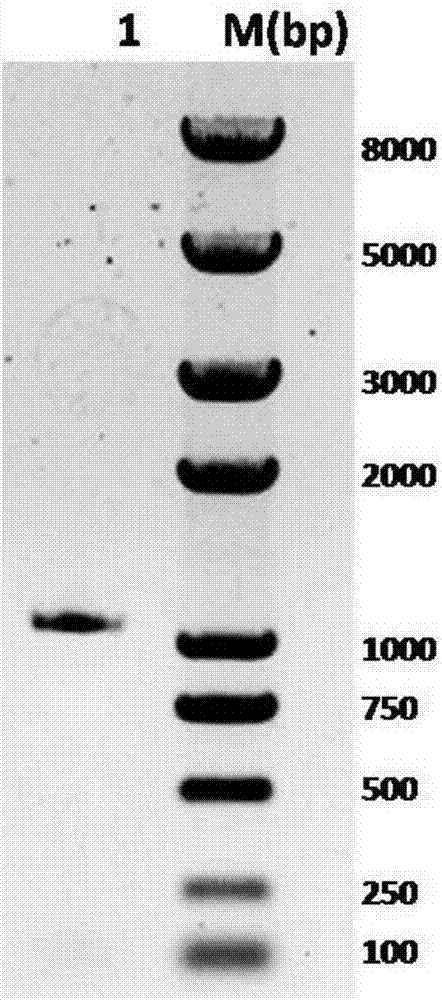
[0039] According to the principle of homologous recombination of In-Fusion (Clontech Company), with the correctly sequenced plasmid pEASY-OcUGE2 as a template, pET28aUGE2 (5'-CGCGGATCCGAATTCATGGCGGGTTTGAACA-3', SEQ ID NO.8) and pET28aUGE2 (5'-TGCGGCCGCAAGCTTCTAATTATTCACTAGT-3 ', SEQ ID NO.9) as primers, PCR was carried out through the following procedures and systems, 50 μL system containing 10×PCR buffer 5 μL, 2mM dNTPs 5 μL, 25mM MgSO4 3 μL, 10 μM primers pET28aUGE2 and RETduetUGE2 each 1.5 μL, KOD-Plus -Neo 1 μL, plasmid 1 μL, ddH2O make up. PCR program: pre-denaturation at 94°C for 5 min; denaturation at 98°C for 30 s, renaturation at 63°C for 45 s, extension at 68°C for 120 s, a total of 30 cycles; extension at 68°C for 7 min, and incubation at 4°C. A 1kb OcUGE2 gene was amplified. The gene was connected to pET-28a treated with EcoRI and HindIII double enzymes by In-Fusion method, the recombinant product was tr...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Concentration | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com