Group of nucleic acid aptamers for isozymes of creatine kinases and application of nucleic acid aptamers
A nucleic acid aptamer, creatine kinase technology, applied in genetic engineering, recombinant DNA technology, biochemical equipment and methods, etc., can solve problems such as unreported, and achieve the effect of high affinity and high binding specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0062] 1. Selection and synthesis of single-stranded DNA library
[0063] The following 5 kinds of DNA were synthesized by Sangon Bioengineering (Shanghai) Co., Ltd.: a DNA library containing 81 bases, its nucleic acid sequence is: ATCCAGAGTGACGCAGCA(N 45)TGGACACGGTGGCTTAGT; "P1 biotin upstream": ATCCAGAGTGACGCAGCA; "P2 "Biotin downstream": ACTAAGCCACCGTGTCCA; "P3 upstream": ATCCAGAGGTGACGCAGCA; "P4 downstream": ACTAAGCCACCGTGTCCA. 1 OD DNA per tube, when needed, centrifuge at 12,000 rpm for 0.5 min before opening the cap; follow the instructions on the tube wall to prepare a 100 μM storage solution.
[0064] 2. Tosyl magnetic beads bound to creatine kinase isoenzyme
[0065] The tosyl magnetic bead (Dynal, 2mL) reagent bottle needs to be vortexed before use. Take 16.5μL (2×10 7 1) magnetic beads, wash twice with Buffer B (0.1M Na-phosphate buffer pH=7.4), add 1mL each time, after adding, vortex, place the 1.5mL centrifuge tube on the magnetic collector, after 1min, When t...
Embodiment 2
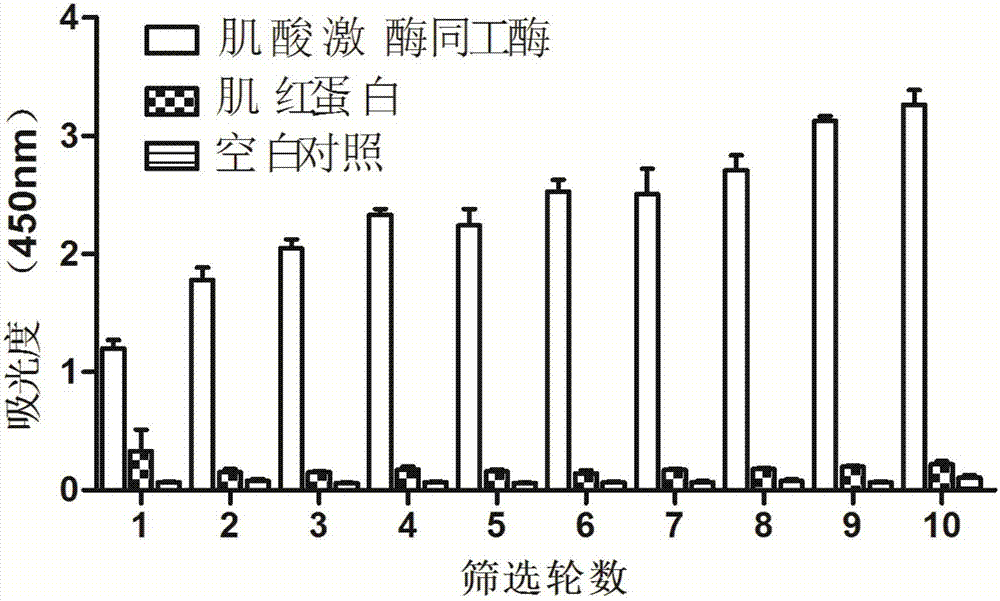
[0117] Get the creatine kinase isoenzyme nucleic acid aptamer described in embodiment 1, carry out the mensuration of dissociation constant respectively, concrete assay method is as follows:
[0118] (1) In a 96-well plate, 100 μL per well of creatine kinase isoenzyme (PROSPEC, 1 mg / 126.5 μL) solution at a concentration of 2000 ng / mL, incubated at 4°C overnight or at 37°C for 2 hours; creatine kinase isoenzyme Coating buffer (1.59gNa 2 CO 3 , 2.93 g NaHCO 3 , dissolved in 1L ultrapure water) for dilution.
[0119] (2) The liquid in the 96-well plate after the treatment in step (1) was emptied and patted dry, and washed twice with 200 μL phosphate Tween buffer, 1 min each time.
[0120] (3) Add 300 μL of blocking solution to each well of the 96-well plate treated in step (2), and incubate at 4°C for 1 hour; Prepared in saline buffer;
[0121] (4) Wash the 96-well plate treated in (3) three times with 200 μL phosphate Tween buffer, 1 min each time;
[0122] (5) Use phospha...
Embodiment 3
[0132] A method for preparing a colloidal gold-labeled A nucleic acid aptamer, the steps are as follows:
[0133] (1) A nucleic acid aptamer with the nucleotide sequence described in SEQ ID No.2, which is modified by sulfhydryl groups and has 20 thymine sequences attached to one end, is added to TE buffer solution (Tris-EDTA buffer) to form solution 1 , adding tris(2-carboxyethyl)phosphine (TCEP) to solution 1, and reacting for 2 hours, the molar ratio of TCEP to the sulfhydryl-modified nucleic acid aptamer of the present invention is 5:1, to obtain solution 2;
[0134] (2) Take 1 mL of colloidal gold solution, centrifuge at 8000 r / min for 30 min, discard the supernatant, add 100 μL of ultrapure water, mix 12 μL of solution 2, shake for 1 min, and overnight to obtain solution 3;
[0135] (3) Add 40 mM NaCl aqueous solution equal to the volume of solution 3 to solution 3 to obtain solution 4 with a concentration of 20 mM, shake for 1 min, and react at room temperature for 8 h; ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com