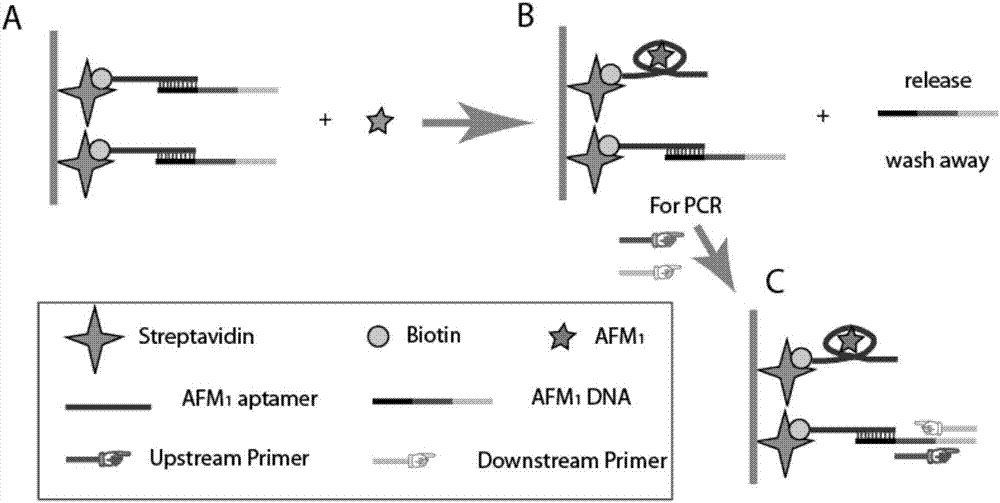
Aptamer for detecting aflatoxin M1, sensor, kit and application thereof
A kind of technology of aflatoxin and nucleic acid aptamer, applied in the direction of biological testing, instruments, measuring devices, etc., can solve the problems of limitation and difficulty in guaranteeing antibody stability, and achieve the effect of lower detection limit, high sensitivity and good selectivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0054] The preparation of embodiment 1 sensor solution
[0055] Mix DNA Sequence A and DNA Sequence B reactions. Wherein, the concentrations of DNA sequence A and DNA sequence B are both 10 nM.
[0056] Wherein, the sequence of DNA sequence A is:
[0057] 5'-GGTGTGACGGATAATCTGGTTTAGCTACGCCTTCCCCGTGGCGATGTTTCTTAGCGCCTTAC-3'.
[0058] The sequence of DNA sequence B is:
[0059] 5'-ATCCGTCACACCTGCTCTGACGCTGGGGTCGACCCG-Biotin-3'.
[0060] Prepare PCR system solution:
[0061] PCR primer 1, PCR primer 2, Premix Ex Mix with ROX Reference Dye II (50×) reaction solution; wherein, the concentrations of PCR primer 1 and PCR primer 2 are both 10 μM.
[0062] Wherein, the sequence of PCR primer 1 is:
[0063] 5'-GTAAGGCGCTAAGAAACATCG-3'.
[0064] The sequence of PCR primer 2 is:
[0065] 5'-AATCTGGTTTAGCTACGCCTTC-3'.
Embodiment 2
[0066] Embodiment 2 makes standard curve
[0067] Treat the PCR tube with 50 μL of 0.8% glutaraldehyde solution at 37°C for 5 hours; after washing with ultrapure water three times, add 50 μL of streptavidin dissolved in 0.01M carbonate buffer and incubate for 2 hours (37°C);
[0068] After washing twice with phosphate buffer, the aptamer and complementary DNA 1:1 (v / v) were thoroughly mixed, and 50 μL of the mixture was added to each tube and incubated for 1 h (37°C);
[0069] Hybridization buffer was washed three times, and 50 μl of aflatoxin M with different concentration gradients (concentrations were 0.1ng / L, 1ng / L, 0.01μg / L, 0.1μg / L, 1μg / L) 1 Standard;
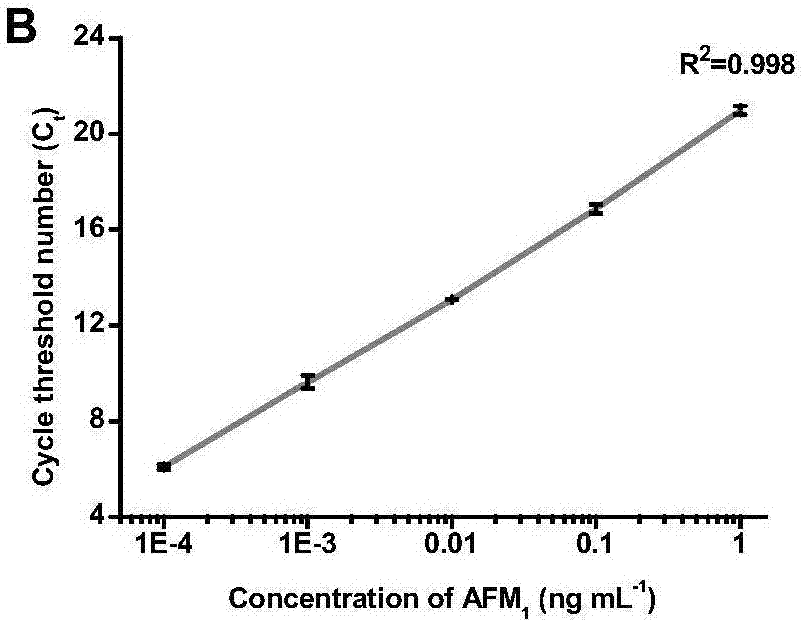
[0070] After washing with Tris buffer three times, add the PCR system solution, detect the fluorescent signal by quantitative PCR, and make a standard curve, see figure 2 .
Embodiment 3
[0071] Aflatoxin M in the detection sample of embodiment 3 1 Content
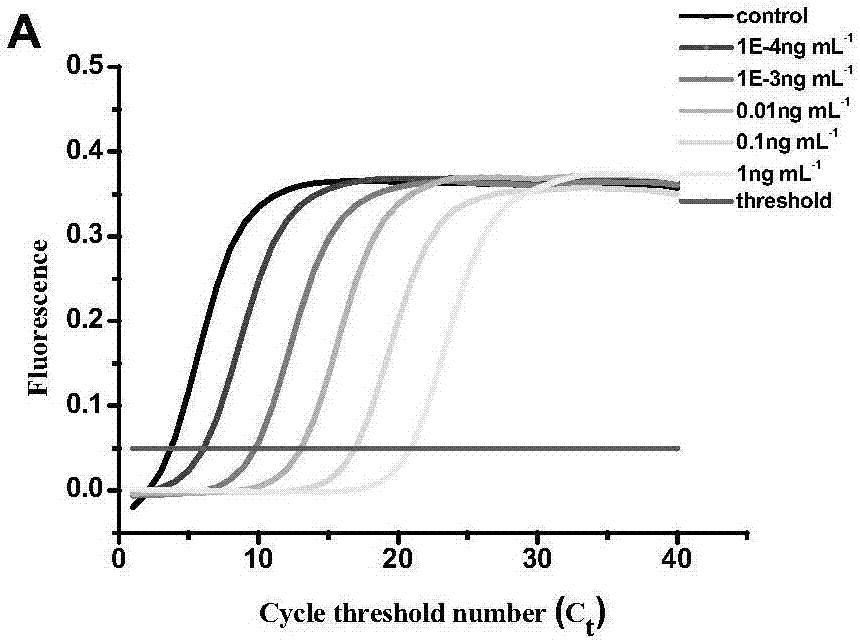
[0072] Weigh 0.5g of the tested sample, add 2.5mL 0.05ng mL -1 Aflatoxin M 1 The standard solution was mixed well with it, and then 2.5mL of 70% methanol was added. The mixture was vortexed for 5 min and then centrifuged at 10000 g for 10 min. The supernatant was collected and concentrated to 0.5 mL by nitrogen blowing. Add 2mL of 5% methanol to redissolve; quantitative PCR detects the fluorescent signal.
[0073] Quantitative PCR detection of aflatoxin M 1 For the amplification curve see image 3 , according to the standard curve measured in Example 2, determine the aflatoxin M in the detection sample 1 The content is 0.045ng mL -1 .
[0074] SEQUENCE LISTING
[0075] Beijing Institute of Animal Husbandry and Veterinary Medicine, Chinese Academy of Agricultural Sciences
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com