Application of PSA3 (photosystem I assembly 3) protein in assembly of PSI (photosystem I) and maintaining of PSI stability
A protein and stability technology, applied in the application field of PSA3 protein in participating in the assembly of PSI and maintaining the stability of PSI, can solve the problems of poorly understood biogenesis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0092] Example 1. Function discovery of PSA3 protein
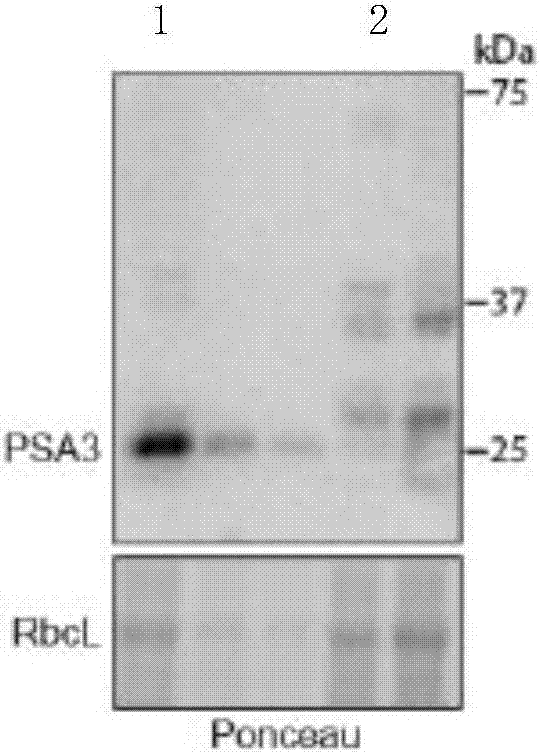
[0093] The inventors of the present invention constructed a Mu-transposon-induced maize mutant library, from which the mutant strains (plants with yellowing phenotype) that could not undergo photosynthesis were screened, and the Mu-transposon insertion and phenotype were identified by a deep sequencing method Co-isolated, several insertion mutations were identified. A mutant strain has an insertion site near the start codon of a specific gene, and the mutant strain is named mutant strain A. Another mutant strain has an insertion site in the third exon of the same specific gene, and this mutant strain is named mutant strain B. Mutants A and B lack the PSI response center protein (PsaA protein), but the levels of PSII core subunit, cytochrome b6f complex, ATP synthase and Rubisco are basically the same as those of the wild plants. The leaves of the plants show increased chlorophyll fluorescence. When the energy of the seed is...
Embodiment 2
[0095] Example 2. Comparison of mutant corn and wild-type corn
[0096] Using the seed embryo of maize HiII as the receptor, the Zm-PSA3 gene was knocked out using CAS9 technology to obtain a mutant maize, which was named Zm-psa3 mutant. After whole-genome sequencing, the Zm-psa3 mutant differs from maize HiII only in that the Zm-PSA3 gene in one chromosome loses its coding function, and the Zm-PSA3 gene in the other chromosome is the same as the receptor.
[0097] The plants to be tested were the homozygous progeny of maize HiII and Zm-psa3 mutants (ie, the Zm-PSA3 genes in both chromosomes have lost their coding function).
[0098] 1. Phenotype identification
[0099] See the phenotype of the plant to be tested at the three-leaf stage figure 1 . figure 1 In the middle, the plant on the left is maize HiII, and the plant on the right is the homozygous progeny of the Zm-psa3 mutant. Compared with maize HiII, the homozygous progeny of Zm-psa3 mutant grows slowly.
[0100] Use mini-PAM...
Embodiment 3
[0105] Example 3 Comparison of wild-type Arabidopsis, mutant Arabidopsis, and complementing strains of Arabidopsis
[0106] The literature mentioning the pCAMBIA1300-flag vector (the corresponding name in the literature is "a modifiedpCAMBIA1300 containing either the Myc or Flag coding sequence"): Zhao Y, Xie S, Li X, Wang C, Chen Z, Lai J, Gong Z. . REPRESSOR OF SILENCING5 Encodes a Member of the Small Heat Shock Protein Family and Is Required for DNA Demethylation in Arabidopsis. Plant Cell. 2014 Jun; 26(6): 2660-2675..
[0107] 1. Obtainment of replenishment strains
[0108] 1. Insert sequence 4 from the 5'end of the double-stranded DNA molecule represented by nucleotides 1-831 from the 5'end between the XbaI and SalI restriction sites of the pCAMBIA1300-flag vector to obtain a recombinant plasmid. According to the sequencing results, the structure of the recombinant plasmid is described as follows: between the XbaI and SalI restriction sites of the pCAMBIA1300-flag vector, the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com