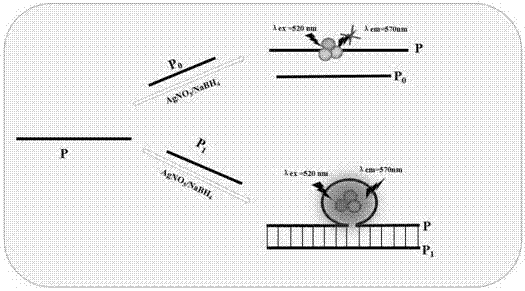
Ag NCs and G-quadruplex/NMM system-based label-free DNA molecular device
A technology of molecular devices and quadruplexes, which is applied in the field of label-free DNA molecular devices, to achieve the effect of selective flexible application and good selectivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
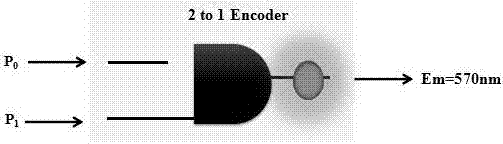
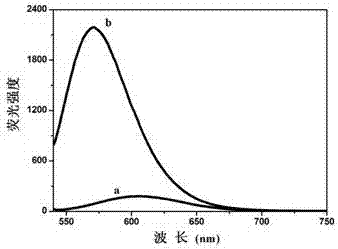
[0089] Such as figure 1 It is the schematic diagram of the 2-to-1 encoder, figure 2 It is the logic circuit diagram of the 2-to-1 encoder, image 3 is the fluorescence intensity curve of AgNCs at different input values of 2-to-1 encoder and Figure 4 It is the 3D histogram of the normalized value of the fluorescence intensity of AgNCs at different input values of the 2-to-1 encoder:
[0090] Design and determination of a 2-to-1 encoder fluorescence system:
[0091] Construction of 2-to-1 encoder:
[0092] Based on single-stranded P-DNA, prepare P-DNA (10μL, 100μmol / L), Mg(AC) 2 (10μL, 100mmol / L) and PBS buffer (950μL, 20mmol / L, PH=7.00) solution, divided into:
[0093] Add P0-DNA (10 μL, 100 μmol / L) to the solution of group a;
[0094] Add P1-DNA (10 μL, 100 μmol / L) to the solution of group b;
[0095] Add AgNO after incubation at 37°C for half an hour 3 (10μL, 0.6mmol / L), magnetically stirred for 2 minutes, incubated at 4°C in the dark for 30 minutes, and quickly...
Embodiment 2
[0103] Such as Figure 5 is the schematic diagram of the 4-to-2 encoder, Figure 6 It is the logic circuit diagram of the 4-to-2 encoder, Figure 7 is the fluorescence intensity curve of AgNCs and NMM at different input values of 4-to-2 encoder and Figure 8 It is the 3D histogram of the normalized value of the fluorescence intensity of AgNCs and NMM at different input values of the 4-to-2 encoder:
[0104] Design and measurement of 4-to-2 encoder fluorescence system:
[0105] Based on single-stranded HS-DNA, the
[0106] a sample solution: P0-DNA (10 μL, 100 μmol / L) solution + HS-DNA (10 μL, 100 μmol / L)) solution;
[0107] b sample solution P1-DNA (10 μL, 100 μmol / L) + HS-DNA (10 μL, 100 μmol / L)) solution;
[0108] c sample solution P2-DNA (10 μL, 100 μmol / L) + HS-DNA (10 μL, 100 μmol / L)) solution;
[0109] d sample solution P3-DNA (10 μL, 100 μmol / L) solution + HS-DNA (10 μL, 100 μmol / L)) solution;
[0110] Add Mg(AC) to the four sample solutions of the above solu...
Embodiment 3
[0126] Such as Figure 9 It is the schematic diagram of the 1-to-2 decoder, Figure 10 is the fluorescence intensity curve of AgNCs and NMM at different input values of the 1-to-2 decoder and Figure 11 It is the 3D histogram of the normalized value of the fluorescence intensity of AgNCs and NMM at different input values of the 1-to-2 decoder:
[0127] Design and determination of 1-to-2 decoder fluorescence system:
[0128] Ag NCs formed with P / StrB1 and NMM / K with strong emission at 570nm + The system is based on:
[0129] In group a test tube: add P-DNA (10 μL, 100 μmol / L), StrB1-DNA (10 μL, 100 μmol / L), Mg(AC) 2 (10μL, 100mmol / L) and PBS buffer solution (950μL, PH=7.00), add ultrapure water (10μL);
[0130] b Two groups of test tubes: add P-DNA (10 μL, 100 μmmol / L), StrB1-DNA (10 μL, 100 μmmol / L), Mg(AC) 2 (10μL, 100mmol / L) and PBS buffer solution (950μL, PH=7.00), add StrB2-DNA (10μL, 100μmmol / L);
[0131] Add AgNO respectively after incubating at 37°C for 30 mi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com