Plant pollen-specific promoter PSP1 and application thereof
A pollen-specific, plant pollen technology, applied in the fields of molecular biology and genetic engineering, can solve problems such as affecting plant growth and development, poor control of the spatiotemporal specificity of target gene expression, and insignificant improvement effects, and achieves broad application prospects and agricultural Economic value, effect of avoiding biosecurity problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] Example 1. Cloning of rice pollen-specific promoter PSP1
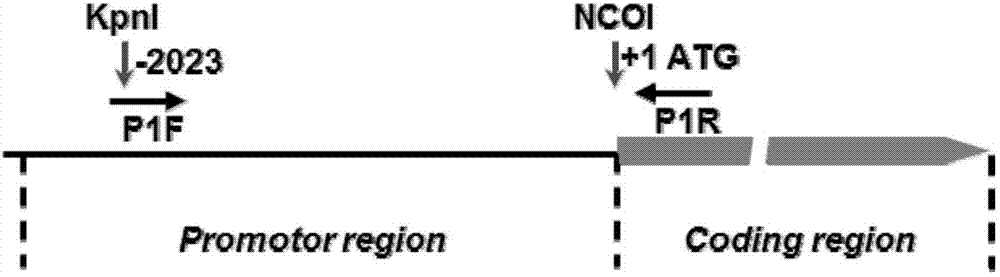
[0039] According to the whole genome sequence of rice variety Nipponbare (Oryza sativa L cv. Nipponbare) provided by NCBI, design specific amplification primers for PSP1 sequence, and add Specific enzyme cutting site ( figure 1 ). The specifically designed primers are: a KpnI restriction site (GGTACC) is added to the 5' end of the forward primer (P1F), and a NcoI restriction site (CCATGG) is added to the 5' end of the reverse primer (P1R). The primer sequences are as follows:
[0040] P1F forward primer: 5'-CGGggtaccAACCAAACCTGTGGCTGC-3'KpnI
[0041] P1R reverse primer: 5'-CATGccatggCGCTCTGTGACGCCAT-3'NcoI
[0042] Then the genomic DNA of the rice variety Nipponbare was extracted, and using the Nipponbare genomic DNA as a template, the primers (P1F and P1R) designed above were used to amplify the promoter PSP1 sequence, and the following amplification procedures were used: pre-denaturation at 94°C for 4 minu...
Embodiment 2
[0043] Example 2. Construction of a plant expression vector driven by the promoter PSP1
[0044] The positive clone obtained in Implementation 1 and identified by sequencing was subjected to plasmid extraction, and the extracted plasmid was double digested with KpnI and NcoI, and the PSP1 promoter fragment was recovered. At the same time, pCambia1305 was linearized by double digestion with KpnI and NcoI, and the pCambia1305 backbone was recovered. The PSP1 promoter fragment recovered after digestion was ligated with the pCambia1305 backbone recovered after digestion with T4 ligase (purchased from NEB Company), Obtain the plant expression vector PSP1-GUS-pCambia1305 ( image 3 ), the PSP1-GUS-pCambia1305 expression vector was transformed into Agrobacterium tumefaciens EHA105 by electric shock transformation method.
Embodiment 3
[0045] Example 3. Transforming rice with the expression vector of the GUS gene driven by the promoter PSP1
[0046] Using the Agrobacterium-mediated method of rice mature embryo callus dissemination transformation, the recombinant expression vector PSP1-GUS-pCambia1305 was transferred into rice mature embryos, and the transformation method was as follows: (1) induction of rice mature embryo callus: mature The Nipponbare seeds were shelled, then surface-sterilized with 75% alcohol for 2 minutes, then soaked in 30% NaClO solution for 30 minutes, and repeated once, and then washed 4-5 times with sterilized water. The seeds were then placed on the induction medium and cultured, and cultured in the dark at 26 degrees to induce callus for transformation ( Figure 4 A). (2) Co-culture of rice callus and Agrobacterium: activate, enrich and resuspend the EHA105 strain identified in Example 2 containing the PSP1-GUS-pCambia1305 expression vector, and adjust OD600=0.3-0.5. Collect the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com