Plagiochasma appendiculatum flavone hexahydroxy o-methyltransferase as well as encoding gene and application thereof
A technology of hydroxyoxymethyl and coding genes, which is applied in the six-position hydroxyoxymethyltransferase of the flavonoids of the purple back moss and its coding gene and application field, and can solve the problem of lack of the purple back moss of the blunt scale.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] Example 1. Cloning of expression gene PaF6OMT
[0046] 1.1 CTAB-PVP method to extract total RNA
[0047] (1) Take the fresh plant material of Violet dorsalis, clean it, blot the excess water with filter paper, put it in a mortar and add liquid nitrogen to grind until the material becomes powder.
[0048] (2) Take an appropriate amount of powder into a pre-cooled centrifuge tube, add 800 μl 65°C preheated CTAB-PVP extract, and mix by inverting up and down.
[0049] The above-mentioned CTAB-PVP extraction buffer preparation method is as follows:
[0050] 100mM Tris HCl (pH 8.0), 2% CTAB (w / v), 2% PVP (w / v), 25mM EDTA, 2M NaCl, mercaptoethanol added to 0.2% after autoclaving; solution configuration was treated with DEPC ddH 2 O, after autoclaving for later use.
[0051] (3) Incubate at 65°C for 30 minutes, and mix by inverting every 6-10 minutes during this period.
[0052] (4) Add 600 μl of chloroform after cooling, mix well; centrifuge at 13,000 rpm at 4°C for 10 mi...
Embodiment 2
[0103] Example 2. Gene protein expression and enzyme activity analysis
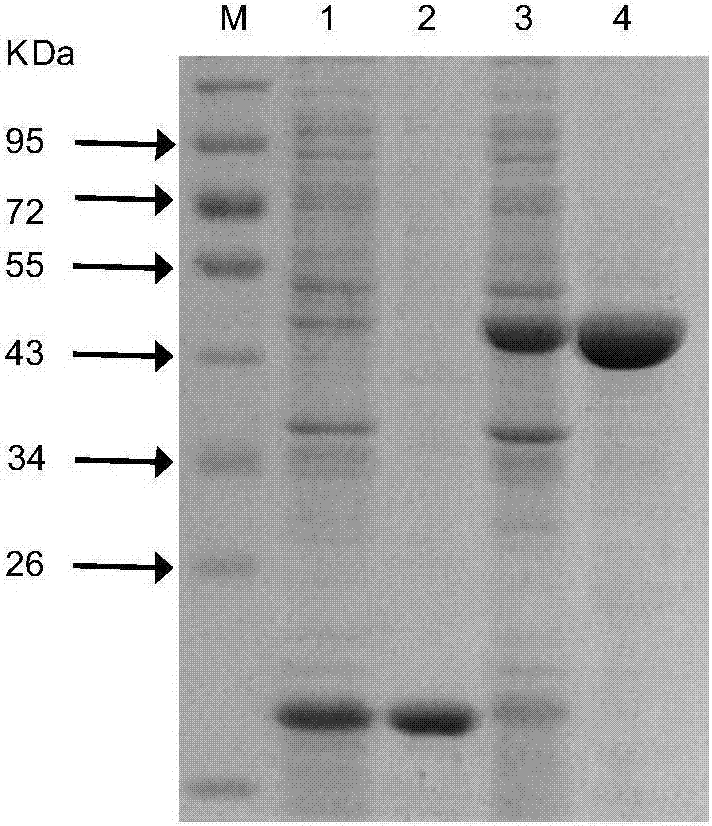
[0104] 2.1 Construction of target gene prokaryotic expression vector
[0105] According to the sequence information of the target gene PaF6OMT and pET32a vector, select the appropriate restriction site to design the primers PaF6OMT-pET32a-F and PaF6OMT-pET32a-R of the prokaryotic expression vector:
[0106] PaF6OMT-pET32a-F:CGGGATCCATGGCCCCAGAGGTTTTGTC; (SEQ ID No. 5)
[0107] PaF6OMT-pET32a-R: CCCTCGAGCTACTTAAGAGTTGAAATGA; (SEQ ID No. 6)
[0108] Using the pMD19T-PaF6OMT plasmid as a template, the above primers were used to amplify its ORF, the PCR product was electrophoresed and gel recovered, and the vector pET32a and the gel recovered fragment were digested with BamH I and Xho I. The enzyme digestion system is as follows:
[0109]
[0110] Digestion was carried out in a water bath at 37°C for 3 hours. Add 10×Loading buffer to the digested product to terminate the reaction, then perform agarose g...
Embodiment 3
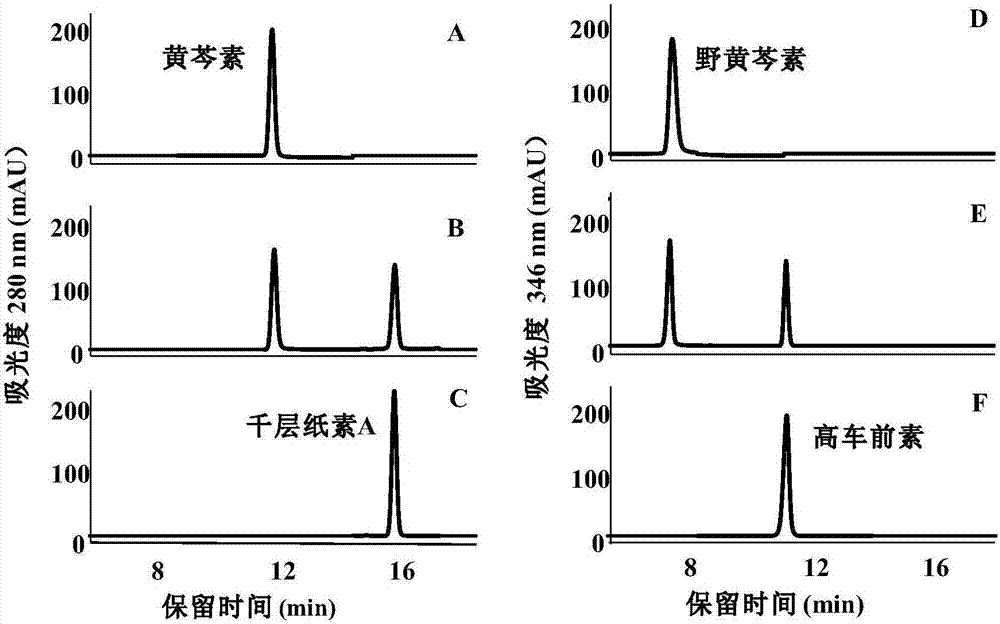
[0170] Embodiment 3.PaF6OMT enzymatic synthesis of homopsylloid and phyllogenin AA
[0171] In the in vitro enzyme function identification experiment, the PaF6OMT protein has a high selectivity for the methylation of 6-OH of baicalein and scutellarein, and the reaction product is single, and the conversion rate of the substrate is high, so it can be used for the enzymatic synthesis of Melaleuca Paper AA and Homo-Psyllogen.
[0172] 3.1 Effect of protein amount on substrate conversion rate
[0173] In order to study the effect of protein amount on the substrate conversion rate, we set the protein amount gradient of 1-14 μg in Tris-HCl (pH 7.5) buffer at 37°C, and the reaction time was 1h. The enzyme activity system is the same as above, wherein SAM and MgCI 2 The amount of added increases proportionally with the increase of protein amount. The effect of protein amount on the substrate conversion rate is as follows: Figure 5A .
[0174] 3.2 Effect of reaction time on subst...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com