A complete set of molecular markers for detecting the chromosome arms of T. villosa and its application
A chromosomal arm, tufted wheat technology, applied in the determination/inspection of microorganisms, biochemical equipment and methods, DNA/RNA fragments, etc. Insufficient utilization, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
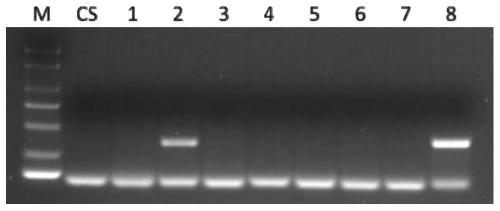
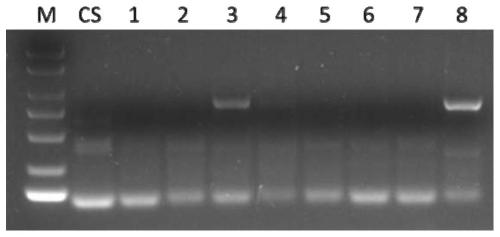
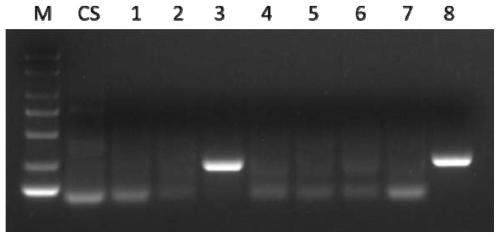
[0106] Example 1. Development of a set of primers for detecting the chromosomal arm of T. villosa
[0107] Trichophyllum for testing: Trichophyllum No.1026, Wheat-Macrophyllum 1V-7V addition line (DA1V-7V), Sclerophyllum amphiploid.
[0108] Transcriptome sequencing: the leaves of T. villosa were taken, RNA was extracted, and transcriptome sequencing was performed to obtain the transcriptome sequence.
[0109] Sequence Alignment: Submit the transcriptome sequence to https: / / urgi.versailles.inra.fr / blast / for comparison, and search for sequences mapped to the genomes of wheat groups 1-7.
[0110] Primer design: According to the sequence compared to the corresponding genome of wheat, primers were designed using primer premier 6, and the primers were synthesized by Sangon Bioengineering (Shanghai) Co., Ltd.
[0111] Preliminary screening of primers: using the genomic DNA of Chinese Spring, Wheat-Tympus villosa 1V-7V addition line (DA1V-7V), and T. villosa No.1026 as templates, P...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com