Probe group for detecting avena plant chromosome, and kit and application
A chromosome and probe group technology, applied in the application field of fluorescent in situ hybridization probes, can solve the problems of indistinguishable genetic relationship between chromosomes, difficult recycling of slices, and affecting the strength of hybridization signals, etc. Easy to slice, easy to prepare, and easy to operate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0060] 1) Preparation of chromosome slide specimens of plants of the genus Avena: Soak the seeds of plants of the genus Avena in double-distilled water, place them in an environment at 4°C for 24 hours, absorb excess water, and culture them at a constant temperature of 22°C until the root tip grows to 1.5 to 2.0 cm, cut 1 to 1.5 cm of root tip tissue, followed by N 2 O treatment for 5 hours, glacial acetic acid treatment for 5 minutes, and then stored in 75% alcohol at -20°C;
[0061] Wash the root tip tissue preserved at low temperature with double distilled water, cut out the root tip meristem 1-2mm, put it in the enzymatic hydrolysis solution at 37°C for 45 minutes, remove the enzymatic solution, wash with double distilled water, 75% alcohol in turn , 95% alcohol, and 100% alcohol to clean the root tip meristem and then dry it at room temperature; add glacial acetic acid to the root tip tissue to make a suspension; drop the suspension on a glass slide and dry it at room tem...
Embodiment 1
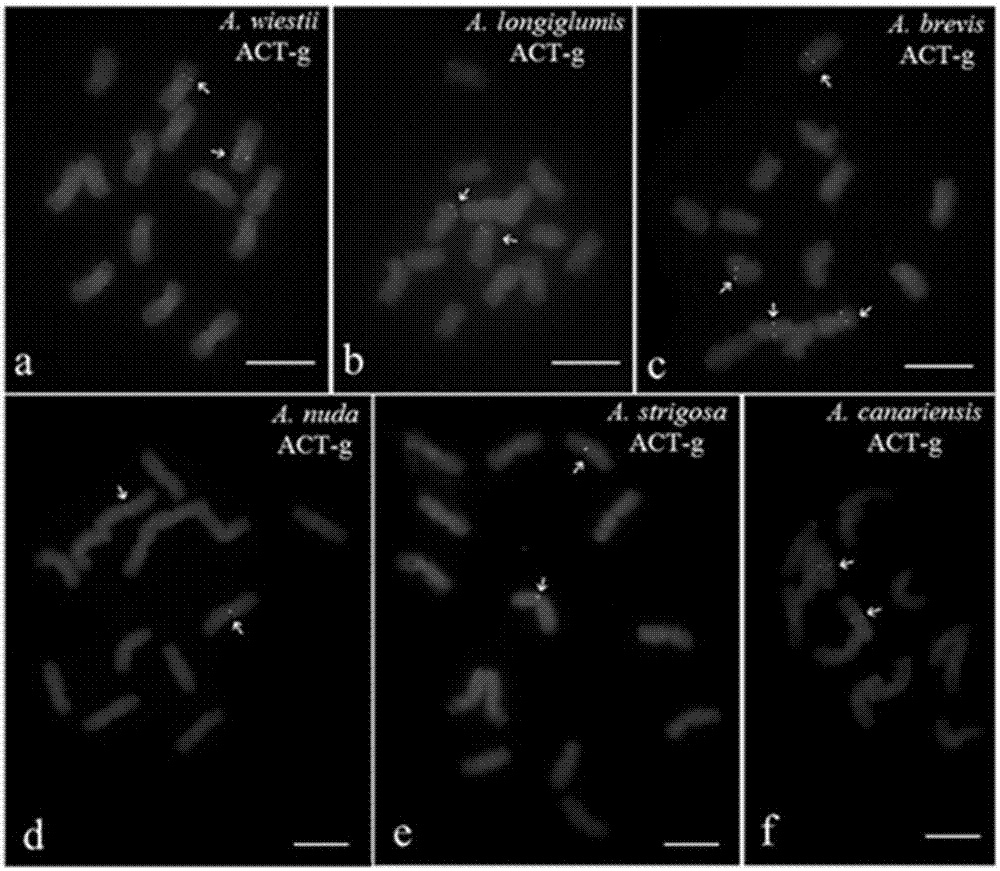
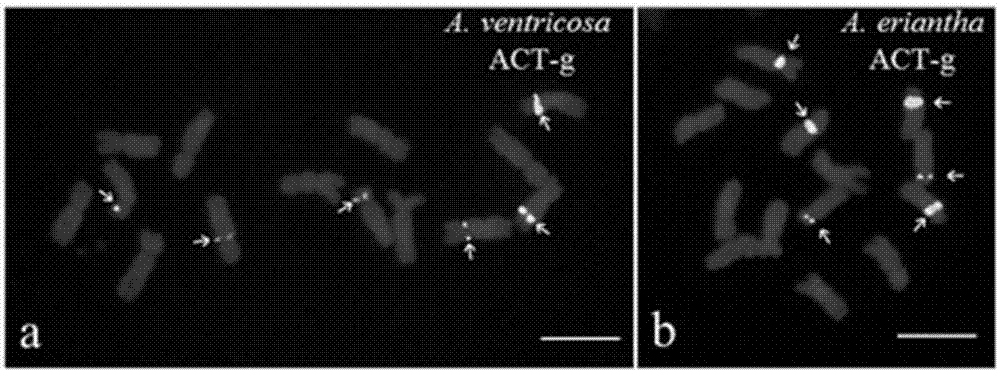
[0068] This embodiment adopts (ACT) in the kit 6 The probes were used to mark six species of Avena diploid plants (AA genome): Avena wiestii, Avena longiglumis, Avena brevis, Avena nuda, Avena strigosa, Avenacanariensis. (ACT) 6 The base sequence of the probe is: 5'-ACTACTACTACTACTACT-3', the 5' end of which is labeled with FAM, showing a green signal, and the amount of the probe is 0.5 μL. The hybridization signals of the above-mentioned 6 diploid plants of the genus Avena can be clearly observed through a fluorescence microscope. In the distribution images of their signal sites, except for the four signals of Avena brevis, the rest are two signals, as shown in figure 1 As shown, the scale bar in the figure is 5 μm, and the arrow points to (ACT) 6 hybridization signal.
Embodiment 2
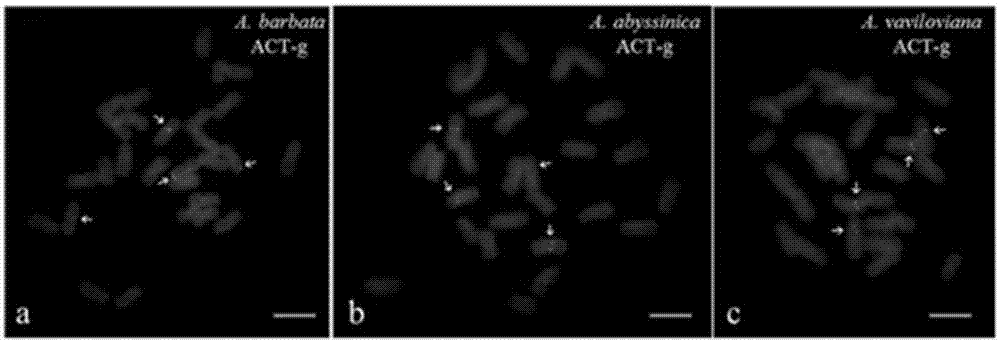
[0070] This embodiment adopts (ACT) in the kit 6 The probes were used to label three tetraploid plants of the genus Avena (AABB genome): Avena barbata, Avena abyssinica, Avena vaviloviana. (ACT) 6 The base sequence of the probe is: 5'-ACTACTACTACTACTACT-3', the 5' end of which is labeled with FAM, showing a green signal, and the amount of the probe is 0.5 μL. The hybridization signals of the above three tetraploid plants of the genus Avena can be clearly observed by fluorescence microscopy, and each species contains four signals, such as figure 2 As shown, the scale bar in the figure is 5 μm, and the arrow points to (ACT) 6 hybridization signal.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com