DNA extraction method
An extraction method and sample technology, applied in the field of DNA extraction, can solve problems such as false negatives, and achieve the effect of simple extraction operation, excellent simplicity and speed, and improved sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Example 1: Adding BSA solution into the PCR system can eliminate the inhibition of PCR by aminated magnetic nanoparticles.
[0038] Preparation of Aminated Magnetic Nanoparticles:
[0039] 1) Fe 3 o 4 Preparation of magnetic nanoparticles: 0.02mol FeCl 3 ·6H 2 O and 0.01mol FeSO 4 ·7H 2 O was dissolved in 150ml of deionized water, heated to 85°C under the protection of nitrogen, and continued to react for 25min, magnetically separated, washed and then vacuum-dried. 2) Preparation of magnetic nanoparticles: 9ml n-hexanol, 37.5mL cyclohexane, 8.85mL Triton 100, 200mg Fe 3 o 4 The nanoparticles were mixed, 0.5 mL tetraethyl orthosilicate was added, and then 0.3 mL ammonia water was added, and the reaction was performed under magnetic stirring at room temperature for 24 hours. Magnetic separation, washing and vacuum drying. 3) Amination modification: 200mg of the above-mentioned Fe 3 o 4 Nanoparticles and 1ml of 3-aminopropyltriethoxysilane were mixed in 30mL of ...
Embodiment 2
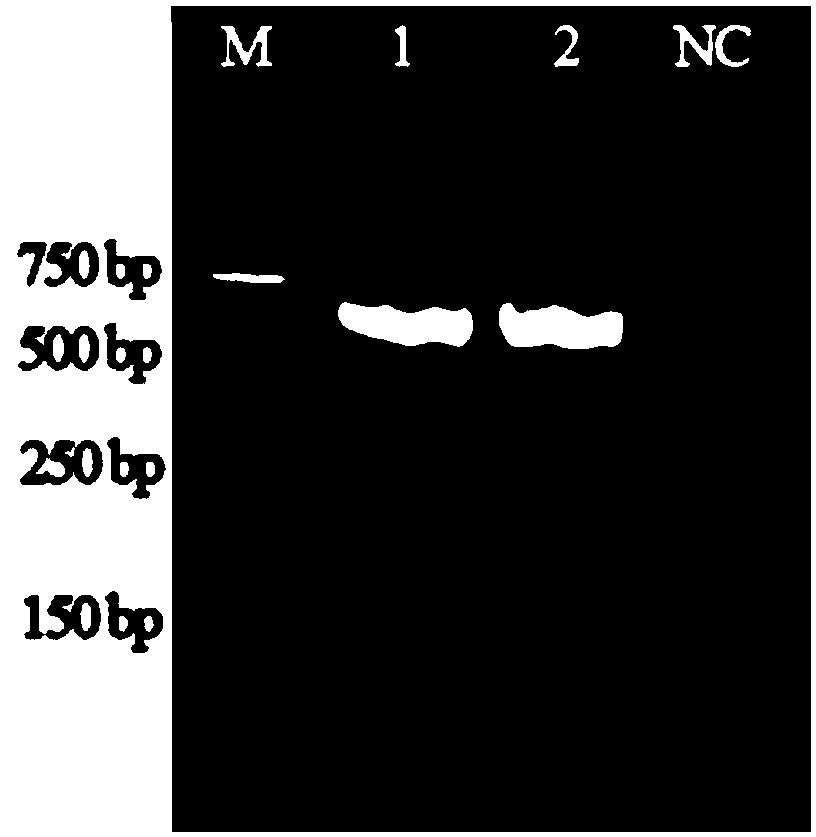
[0041] Example 2: Extract the genomic DNA of the fungal culture, amplify the gene sequence by PCR, and determine whether it is matsutake by sequencing.
[0042]Take a small amount of fungal culture (Shanghai Academy of Agricultural Sciences) and grind it three times with liquid nitrogen, weigh 200 mg sample, add 1000 μL of TE buffer solution (pH=9), add 2 μL of 500 U / mL lysozyme (purchased from Shanghai Yisheng Biotechnology Co., Ltd. Technology Co., Ltd.), incubate at 37°C for 30min. Add 10 μL of Triton X100 (purchased from Shanghai Sinopharm Group) and 50 μL of proteinase K (purchased from Merck) at a concentration of 20 μL / mL, and incubate at 65° C. for 30 min. Centrifuge at 12000rpm for 5min, absorb the supernatant and transfer to a new 1.5mL centrifuge tube, add 40μg of the aminated magnetic nanoparticles prepared in Example 1, and react at 90°C for 10min. After TE was eluted once, the solid component was magnetically separated at the bottom of the small PCR tube, and ad...
Embodiment 3
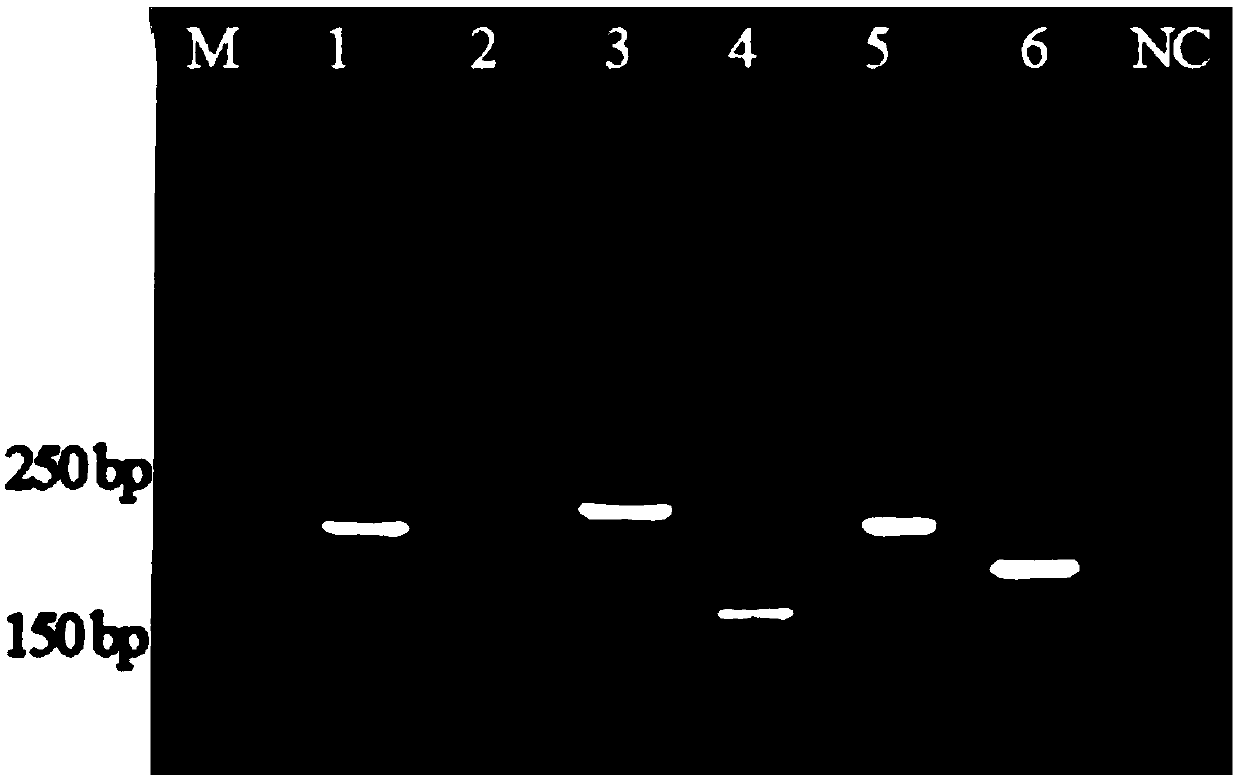
[0044] Example 3: Genomic DNA is extracted from cottonseed hulls, and PCR is used to identify whether it is transgenic cottonseed hulls.
[0045] Take a small amount of cottonseed hulls (from Shanghai Academy of Agricultural Sciences) and grind them three times with liquid nitrogen, weigh 200 mg of the sample, add 1000 μL of TE buffer solution (pH=9), add 10 μL of Triton X100 and 50 μL of protease at a concentration of 20 μL / mL K, 65°C for 30 minutes. Then, treat at 100°C for 10 minutes. Centrifuge at 12000rpm for 5min, draw the supernatant into a new 1.5mL centrifuge tube, add 200μg purchased aminated magnetic beads ( Plus Amine, purchased from Bangslab Company), reacted at 90°C for 10min. After washing once with TE buffer, the solid components were magnetically separated into different PCR small tubes, and added to the PCR system (the system contained 2 μL of 10% BSA solution).
[0046] Amplification primer pairs are: CaMV35S-r / f, NOS-r / f, CpTI-r / f, Cry IA(b)+Cry IA(c)-r...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com