SgRNAs specifically targeting to LAG-3 gene and method for specifically knocking out LAG-3 gene
A LAG-3, specific technology, applied in the field of genetic engineering, can solve the problems of only targeting extracellular targets, not easy to develop antibodies, and diversification of tumor mutations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0084] Example 1 Design and Synthesis of Targeted sgRNA for CRISPR-Cas9 / Cpf1-specific Knockout of Human LAG-3, TIM-3 or PD-1 Gene
[0085] 1. Design of Cas-sgRNA targeting human LAG-3, TIM-3 or PD-1 gene:
[0086] (1) Select the sequence of 5'-N(21)GG or 5'-N(21)AG on the LAG-3, TIM-3 or PD-1 gene.
[0087] (2) The targeting site or cleavage site of the sgRNA on the LAG-3, TIM-3 or PD-1 gene is located in the exon of the gene, which is more likely to cause the deletion of the fragment or the frame-shift mutation, thereby reaching the gene Completely inactivate the purpose.
[0088] (3) The targeting site or cleavage site of the sgRNA on the LAG-3, TIM-3 or PD-1 gene is located on the common exons of different cleavage forms.
[0089](4) Use Blat in the UCSC database or BLAST in the NCBI database to determine whether the target sequence of the sgRNA is unique and reduce potential off-target sites.
[0090] According to the above method, the present invention designed a total...
Embodiment 2
[0123] Example 2 Construction of sgRNA gene editing plasmid
[0124] 1. Construction of Cas9 plasmid:
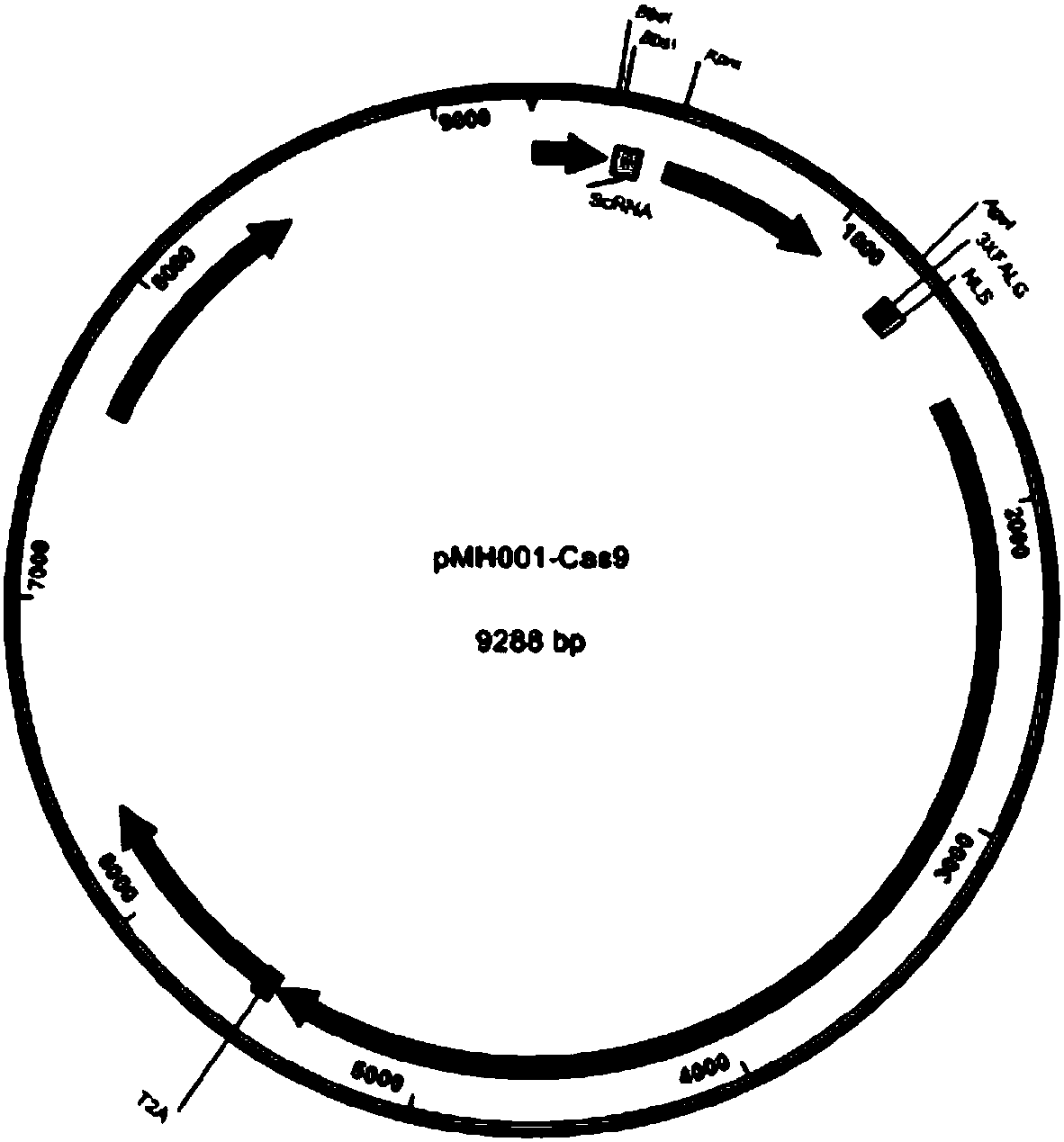
[0125] For the preparation method of plasmid PMH001-Cas9, please refer to the literature: Le Cong et al. Multiplex GenomeEngineering Using CRISPR / Cas Systems Science 339, 819 (2013); (DOI: 10.1126 / science.1231143), please refer to the structure diagram of plasmid PMH001-Cas9 figure 1 , the complete sequence is shown in SEQ ID NO.478.
[0126] 2. Construction of Cpf1 plasmid:
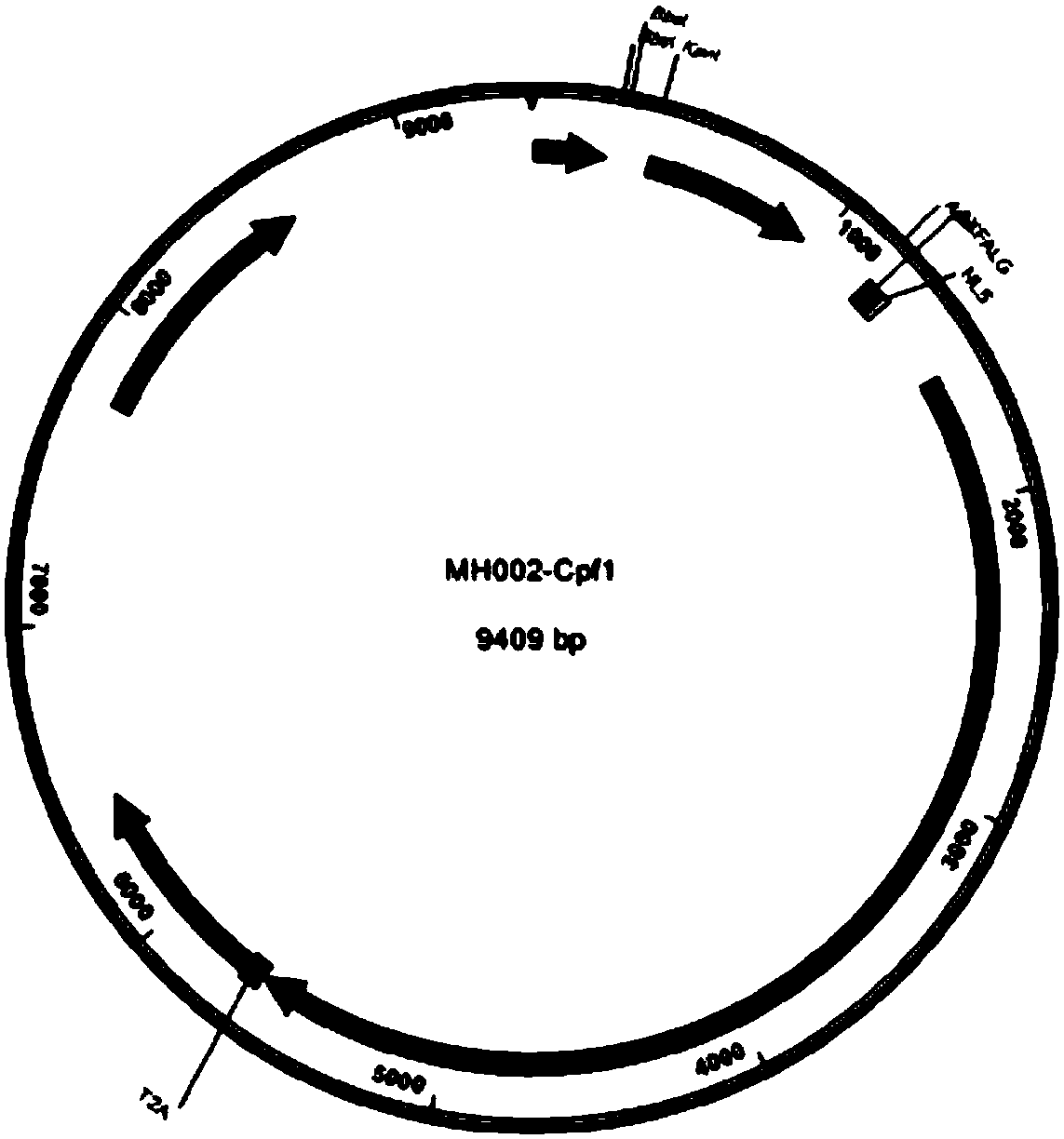
[0127] For the preparation method of plasmid PMH002-Cpf1, please refer to the literature: Zetsche Bet al. figure 2 , the complete sequence is shown in SEQ ID NO.479.
[0128] 3. Construction of single sgRNA gene editing plasmid:
[0129] (1) Linearize the PMH001-Cas9 or PMH002-Cpf1 plasmid prepared in steps 1 and 2 above.
[0130] Enzyme digestion system and conditions are as follows:
[0131] 2μg PMH001-Cas9 or PMH002-Cpf1 (400ng / μl);
[0132] 5μl 10x FastDigest Buffer / FastDigest Green Buffer;...
Embodiment 3
[0154] Example 3 Using CRISPR-Cas9 to specifically knock out the human LAG-3 or TIM-3 gene
[0155] 1. Cell culture and transfection
[0156] (1) HEK293T cells were inoculated and cultured in DMEM medium containing 10% FBS, penicillin (100 U / ml) and streptomycin (100 μg / ml).
[0157] (2) Divide into 12-well plates before transfection, and perform transfection when the density is 60%-80%.
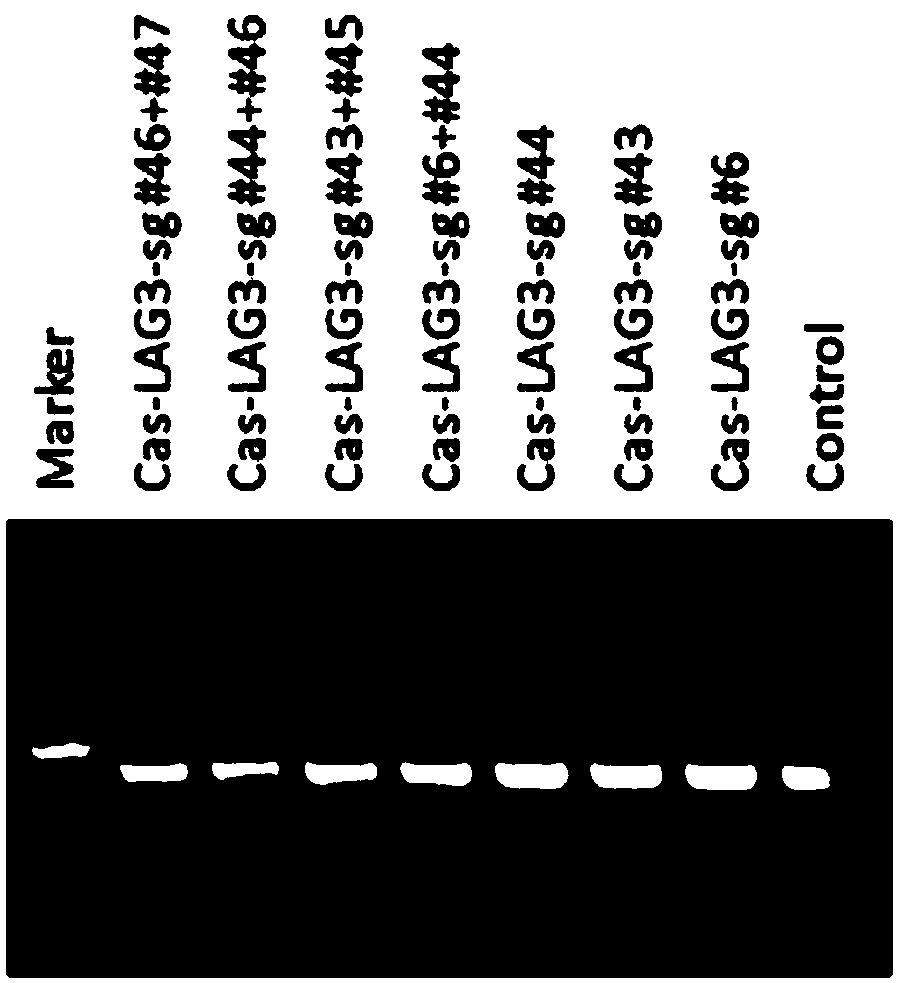
[0158] (3) According to the operation manual of LipofectamineTM2000Transfection Reagent (Invitrogen, 11668-019), transfect 2 μg of the plasmid carrying LAG-3sgRNA or TIM-3sgRNA targeting LAG-3sgRNA or TIM-3sgRNA into each well of cells, change the medium after 6-8 hours, and add Puromycin (Merck, 540411) drug screening, harvested cells after 48 hours.
[0159] Design the experimental group and control group as shown in Table 1:
[0160] Table 1. CRISPR-Cas9 specific knockout of human LAG-3 or TIM-3 gene experimental group design
[0161]
[0162]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com