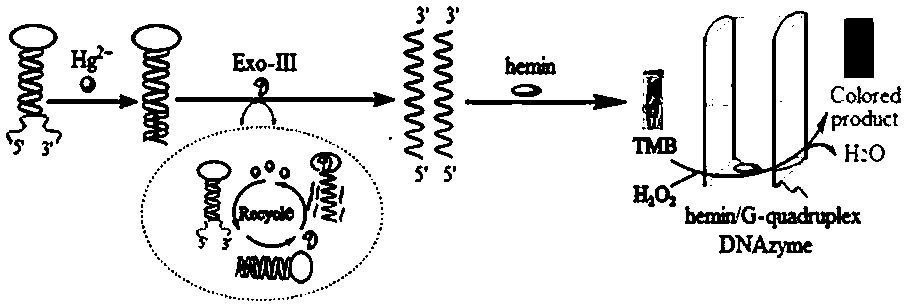
Visual colorimetric sensor for detecting mercury ions
A visual colorimetric and sensor technology, applied in the field of analytical chemistry, can solve the problems of large background interference, high detection cost, complicated operation, etc., and achieve the effect of good reproducibility, good specificity and mild reaction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] 1. Preparation of H-DNA with hairpin structure:
[0035] (1) Prepare Tris-HCl buffer 1 (TB1, pH 7.4), TB1 consists of 20 mM Tris-HCl, 100 mM NaCl and 5mM MgCl 2 composition;
[0036] (2) Dissolve H-DNA in TB1, heat at 90°C for 10 minutes, and then cool to room temperature naturally.
[0037] 2. Preparation of hemin solution:
[0038] (1) Prepare Tris-HCl dilution buffer 2 (TB2, pH 7.4), TB2 consists of 300 mM Tris-HCl, 10mM TCEP, 1 M NaCl, 1mM EDTA and 1 mM MgCl 2 composition;
[0039] (2) First dissolve hemin in dimethyl sulfoxide (DMSO), and then dilute it to the required concentration with Tris-HCl buffer 2.
[0040] 3. Preparation of 3,3',5,5'-tetramethylbenzidine (TMB) solution:
[0041] (1) Prepare PBS buffer (pH 5.5), PBS buffer is made of 10 mM NaH 2 PO 4 , 10 mM Na 2 HPO 4 composition;
[0042] (2) First dissolve the TMB sample in pure ethanol, then add ultrapure water to volume, and dilute to the desired concentration with PBS buffer (pH 5.5).
Embodiment 2
[0044] Pretreatment of actual samples:
[0045] (1) After the environmental water sample was left to stand for a period of time, it was filtered with a 0.22 μm microporous membrane, and dilute nitric acid was added to the filtrate to make the HNO in the sample solution 3 The final concentration is 0.5%, mix well.
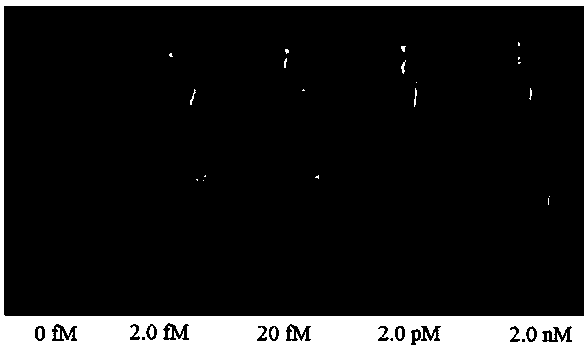
[0046] (2) Divide the tap water sample and the lake water sample into 3 parts, and number them: tap water No. 1, tap water No. 2, tap water No. 3 and lake water No. 1, lake water No. 2, and lake water No. Add standard Hg 2+ The solution was prepared with a spiked concentration of 1.0 fM, and the standard Hg was added to No. 2 tap water and No. 2 lake water. 2+ solution, prepared with a spiked concentration of 1.0 pM; add standard Hg to No. 3 tap water and No. 3 lake water 2+ solution, prepared with a spiked concentration of 1.0 nM.
Embodiment 3
[0048] Testing of actual samples:
[0049] (1) Take 45 μL of H-DNA with a concentration of 2.5 μM and 45 μL of water samples (or water sample spiked solutions with different concentrations), mix well, and add 10 μL of H-DNA with a concentration of 2.5 U / μL to them after 2 min. Exonuclease III (Exo-III), mix well and put it into a K30 economical dry thermostat, and react at 35 °C for 40 min;
[0050] (2) After the above reaction solution is cooled, add 100 μL of hemin (hemin) with a concentration of 7.75 μM to it, mix well and put it in the refrigerator (4°C) for 30 min;
[0051] (3) Add 100 μL of 5.0 mM 3,3',5,5'-tetramethylbenzidine (TMB) and 100 μL of 10.0 mM H 2 o 2 , and observe the color change of the solution after mixing for 1 min.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com