Mutant immunoglobulin kappa chain variable region-binding peptide
An immunoglobulin, binding technology, applied in the direction of immunoglobulin, carrier-bound/immobilized peptide, hybrid peptide, etc., to achieve the effect of excellent selective adsorption capacity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0123] Example 1: Preparation of various mutant VL-κ binding peptides of PpL
[0124] (1) Expression plasmid preparation
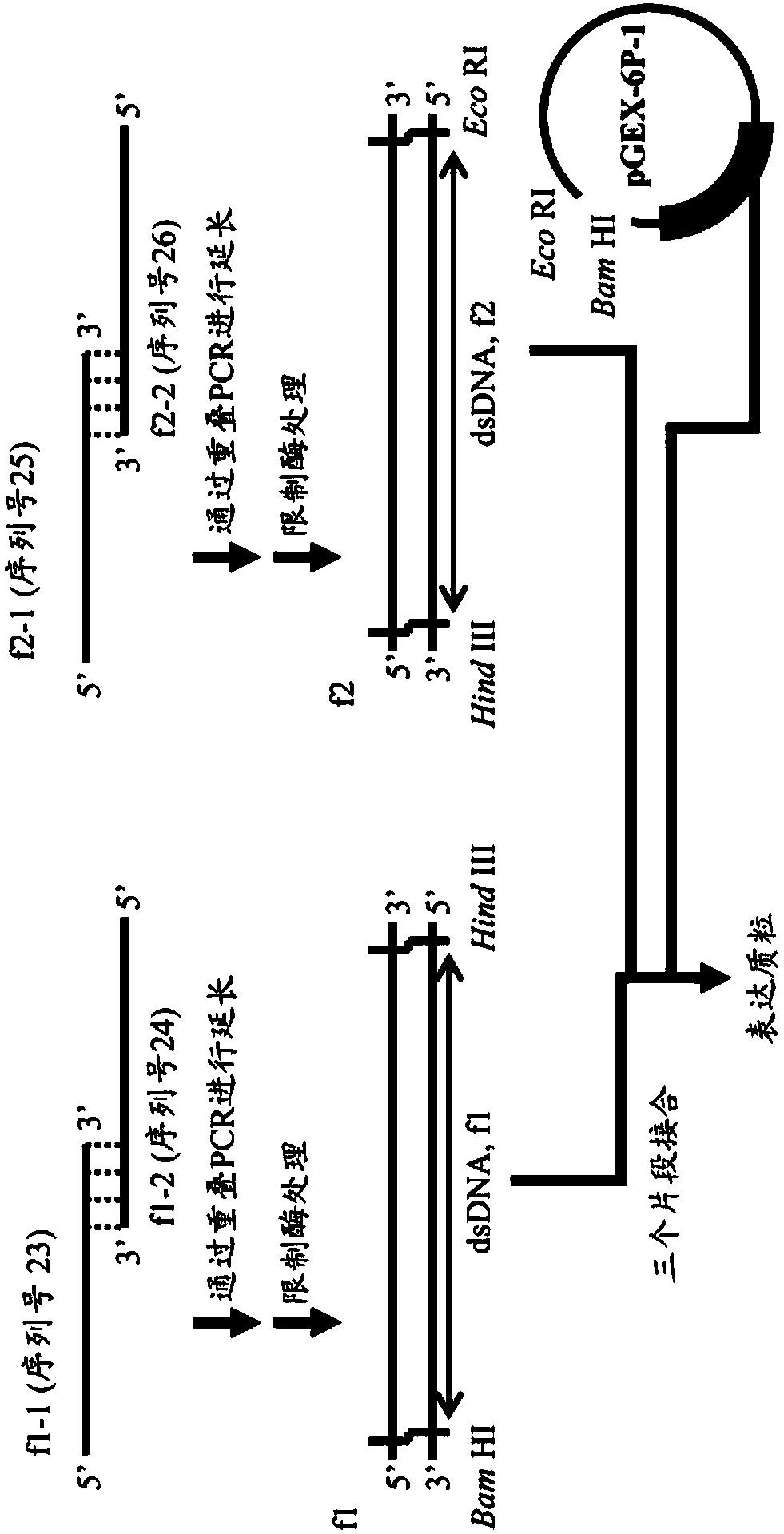
[0125] From the amino acid sequence of LB5t-Wild.1d (SEQ ID NO: 16), the base sequence (SEQ ID NO: 22) encoding the peptide was designed by reverse translation. It should be noted that, based on experimental conditions, the nucleotide sequence was designed to encode an amino acid sequence with Glu-Gln added to the N-terminus and Gly added to the C-terminus. The 1-2 residue addition sequence comes from the B5 domain of wild-type PpL. Then, the preparation method of the expression plasmid is shown in figure 2 . The DNA encoding LB5t-Wild.1d was prepared by ligating two double-stranded DNAs (f1 and f2) having the same restriction enzyme site, and introduced into the multiple cloning site of the expression vector. Actually, preparation of coding DNA and introduction into the vector were carried out simultaneously by ligating two kinds of double-stranded D...
Embodiment 2
[0135] Example 2: Affinity evaluation of various LB5t mutants to aRSV-Fab
[0136] (1) Preparation of Fab fragments from IgG
[0137]As a raw material of humanized monoclonal IgG preparation, it is prepared by fragmenting it into Fab fragment and Fc fragment with papain, and separating and purifying only the Fab fragment. Specifically, an anti-RSV monoclonal IgG (generic name "Palivizumab", product name "Synagis", AbbVie company) preparation whose light chain is formed from a κ chain was dissolved in a buffer for papain digestion ( 0.1M AcOH-AcONa, 2mM EDTA, 1mM cysteine, pH5.5), add "PapainAgarose from papaya latex" papain-immobilized agarose (SIGMA company), and mix it with a rotator, while at 37 ° C Incubate for about 8 hours. From the reaction solution (mixed with Fab fragments and Fc fragments) separated from papain-immobilized agarose, it was recovered in the flow-through fraction by affinity chromatography using a MabSelect SuRe column (GE Healthcare Bioscience). IgG...
Embodiment 3
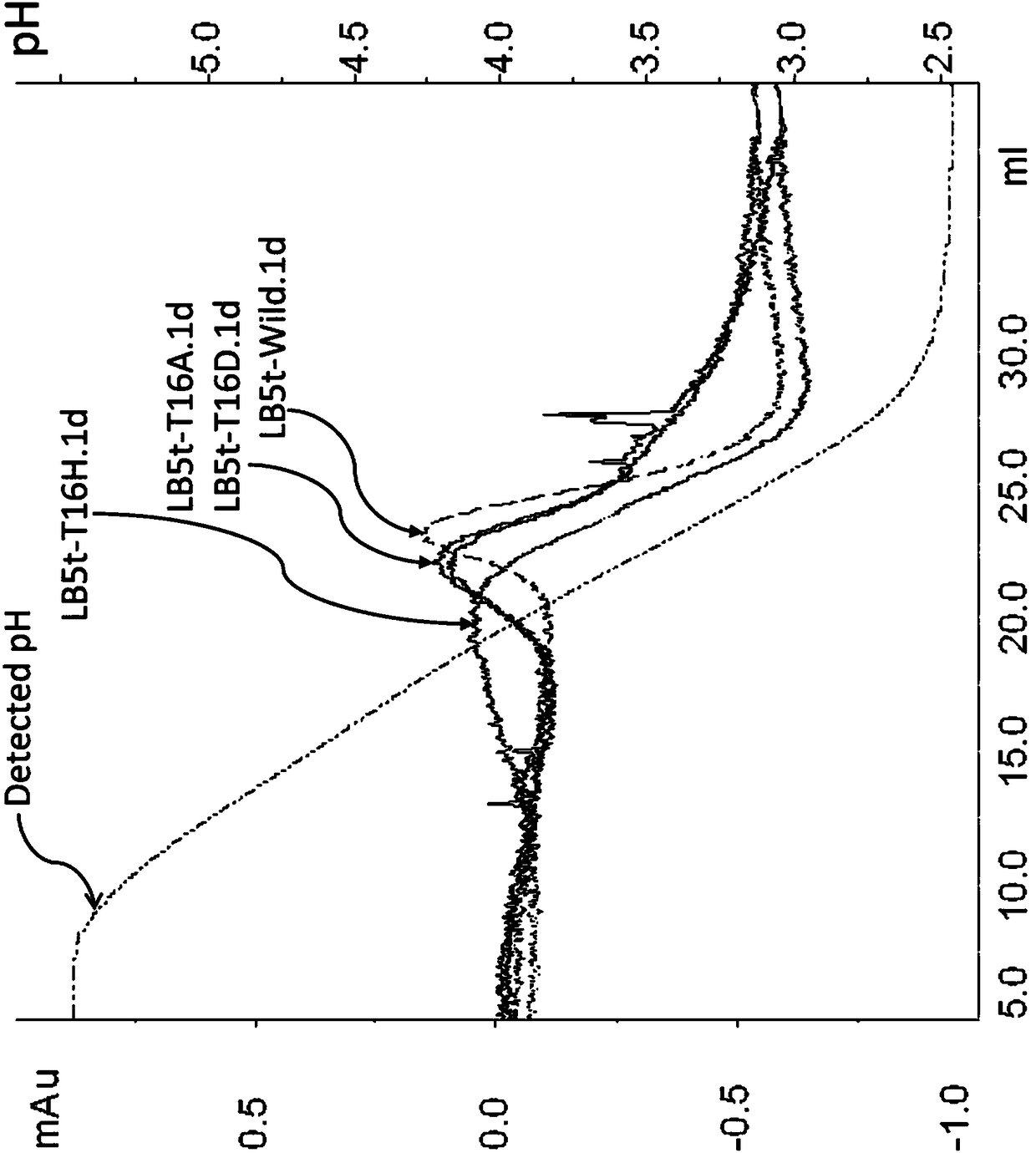
[0143] Example 3: Evaluation of the aRSV-Fab acid dissociation pH of various LB5t mutants
[0144] (1) Preparation of Fab fragment immobilized carrier
[0145] An affinity separation matrix immobilized with the aRSV-Fab obtained in Example 2 was produced using a commercially available coupling column for ligand immobilization in which the coupling target functional group was amino groups.
[0146] As the water-insoluble substrate, 1 mL of a commercially available prepacked column ("Hitrap NHS activated HP" manufactured by GE Healthcare Bioscience) was used. The column uses cross-linked agarose as the matrix, and has introduced active groups for the immobilization of protein ligands whose coupling target functional groups are amino groups. Therefore, the ligands were immobilized according to the product instructions. An operation of passing 1 mM HCl cooled in an ice bath through 2 mL at a flow rate of 1 mL / min was performed three times to remove isopropanol in the column.
[...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com