Real-time fluorescence nucleic acid isothermal amplification detection kit for Cronobacter spp
A technology of Enterobacter zosteri and a kit is applied in the field of biological detection of pathogenic microorganisms, which can solve the problems of false negative detection cost, low sensitivity and false positive experimental results.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0100] Example 1. Design of special primers and probes for real-time fluorescent nucleic acid constant temperature amplification detection of Cronobacter sakazakii (Cronobacter spp)
[0101] Based on personal experience and reading a large number of documents and patents, the present invention selects 23s-ITS2-5S Rrna, 16s-ITS-23S Rrna in the Cronobacter spp gene, 16s rRNA has no secondary structure and is highly conserved The segment is used as the amplified target sequence region. According to the principle of primer probe design, use DNAStar, DNAMAN software and artificially design special primers and probes for real-time fluorescent nucleic acid constant temperature amplification to detect Cronobacter sakazakii (Cronobacter spp) Sequence, the following specific sequence is obtained:
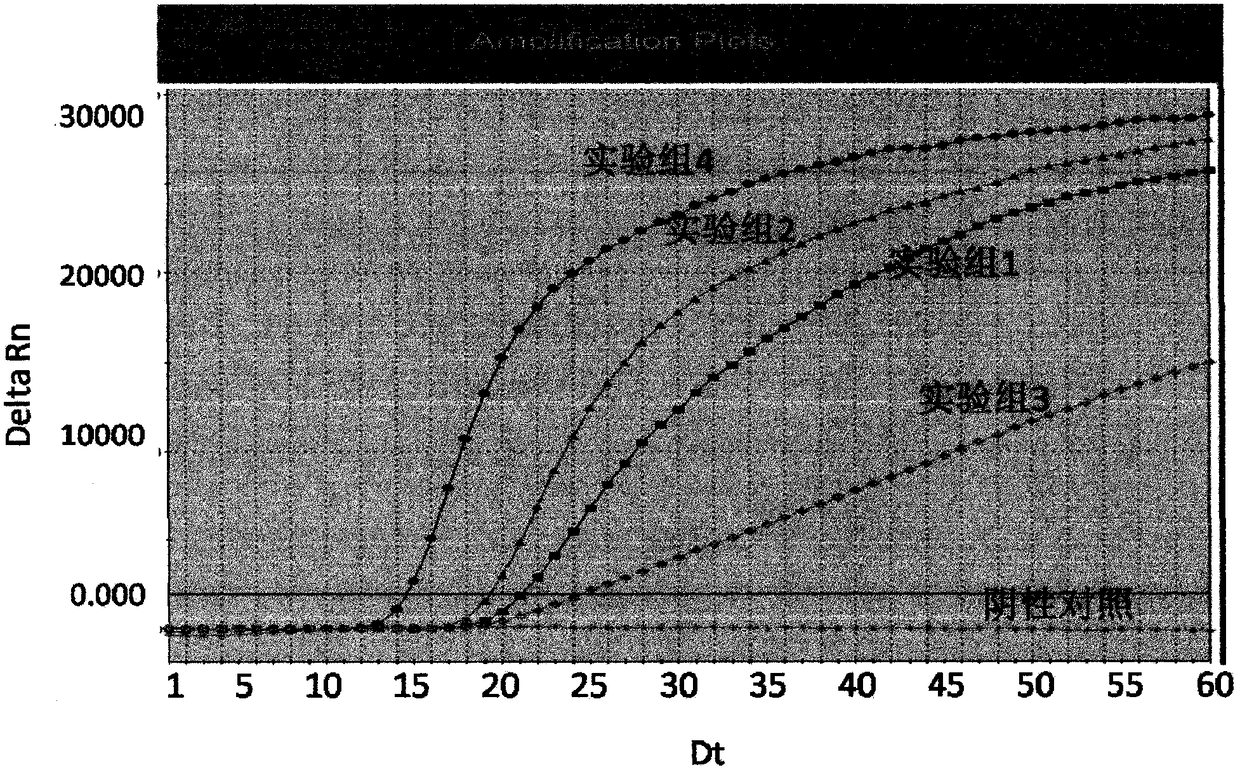
[0102] Experimental group 1:
[0103] nT7: 5'-TTAACCTTACAACGCCAA-3'
[0104] T7: 5'-AATTTATACGACTCACTATAGGGAGACCGCTTCCGCCATGA-3'
[0105] Probe: 5'-CCGCCAGUCUGGCGUGUGGCGG-3'
[0106] Expe...
Embodiment 2
[0125] Example 2. Specific primers and probes for the detection of Cronobacter sakazakii Enterobacter sakazakii (Cronobacter spp) by real-time fluorescent nucleic acid constant temperature amplification
[0126] The primers and probes selected last in Example 1 were selected, and the sequences were put into NCBI BLAST, and the results indicated that this region could detect all subtypes of Enterobacter sakazakii of the genus Cronobacter, namely Cronobacter dublinenesis and Cronobacter dublinenesis subsp lausannensis , Cronobacter malonaticus, Cronobacter muytjensii, Cronobacter sakazakii, Cronobacter turicensis, Cronobacter universalis, Cronobacter genomospecies, Cronobacter condiment.
Embodiment 3
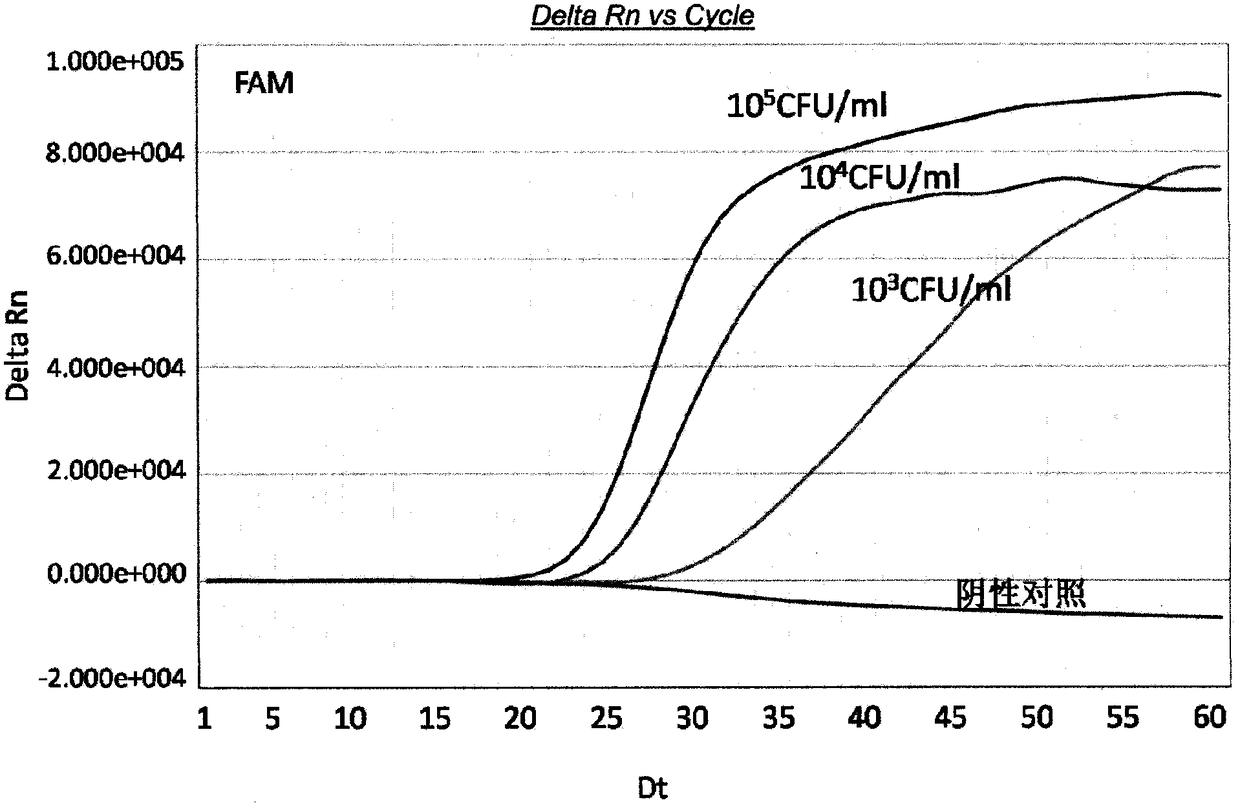
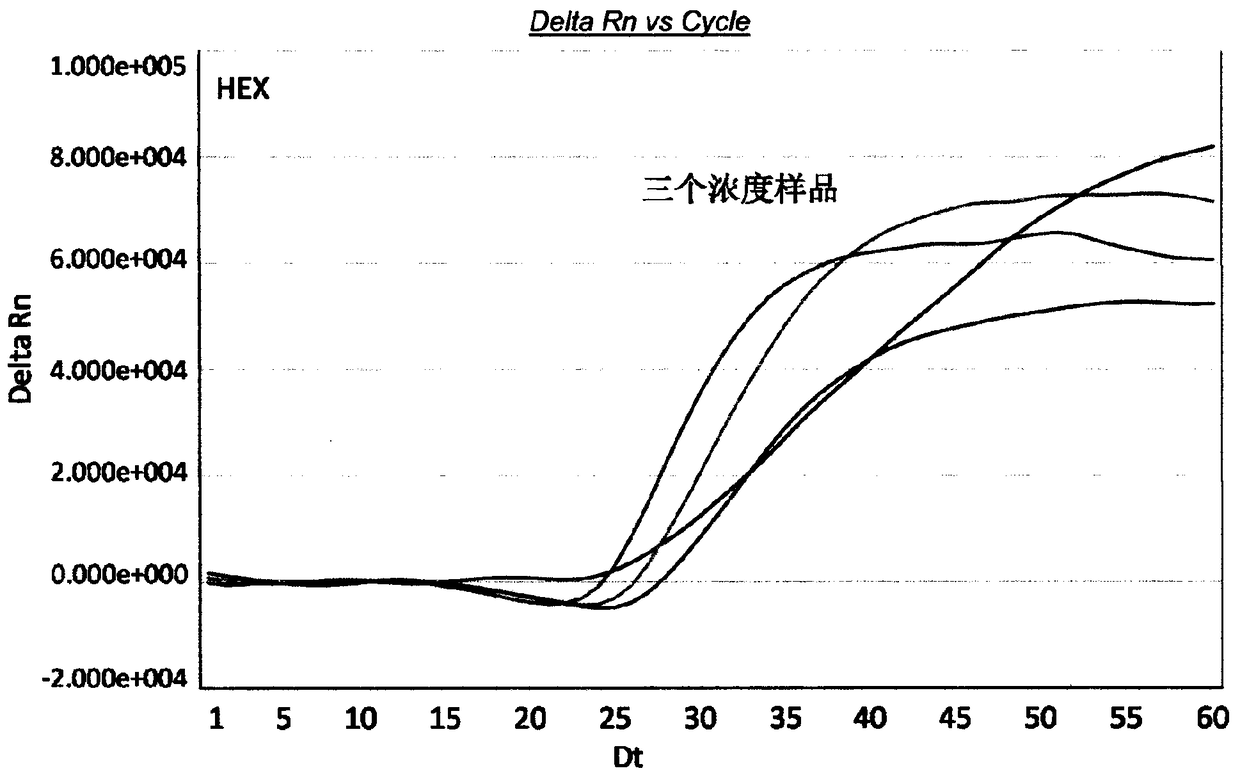
[0127] Example 3. Preparation of a real-time fluorescent nucleic acid constant temperature amplification detection kit for Cronobacter sakazakii (Cronobacter spp)
[0128] Using the special primers and probes provided in Example 1, the real-time fluorescent nucleic acid constant temperature amplification detection kit for Cronobacter sakazakii (Cronobacter spp) of the present invention was obtained. The kit contains components such as capture probe (TCO, Target Capture Oligo), T7 primer, nT7 primer, detection probe, internal standard detection probe, internal standard, M-MLV reverse transcriptase and T7 RNA polymerase .
[0129] The capture probe exists in the nucleic acid extraction solution, the T7 primer, nT7 primer, detection probe, and internal standard detection probe exist in the detection solution, and the M-MLV reverse transcriptase and T7 RNA polymerase exist In the SAT enzyme solution, specifically, the kit is divided into A box (specimen processing unit) stored at...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com