Method for marking germ cells and Sertoli cells of spermatogenic epithelium of turbot in different developmental stages and application
A germ cell and labeling method technology, applied in the field of fish cell labeling, can solve problems such as hindering research, and achieve the effect of high accuracy and simple and easy labeling method.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
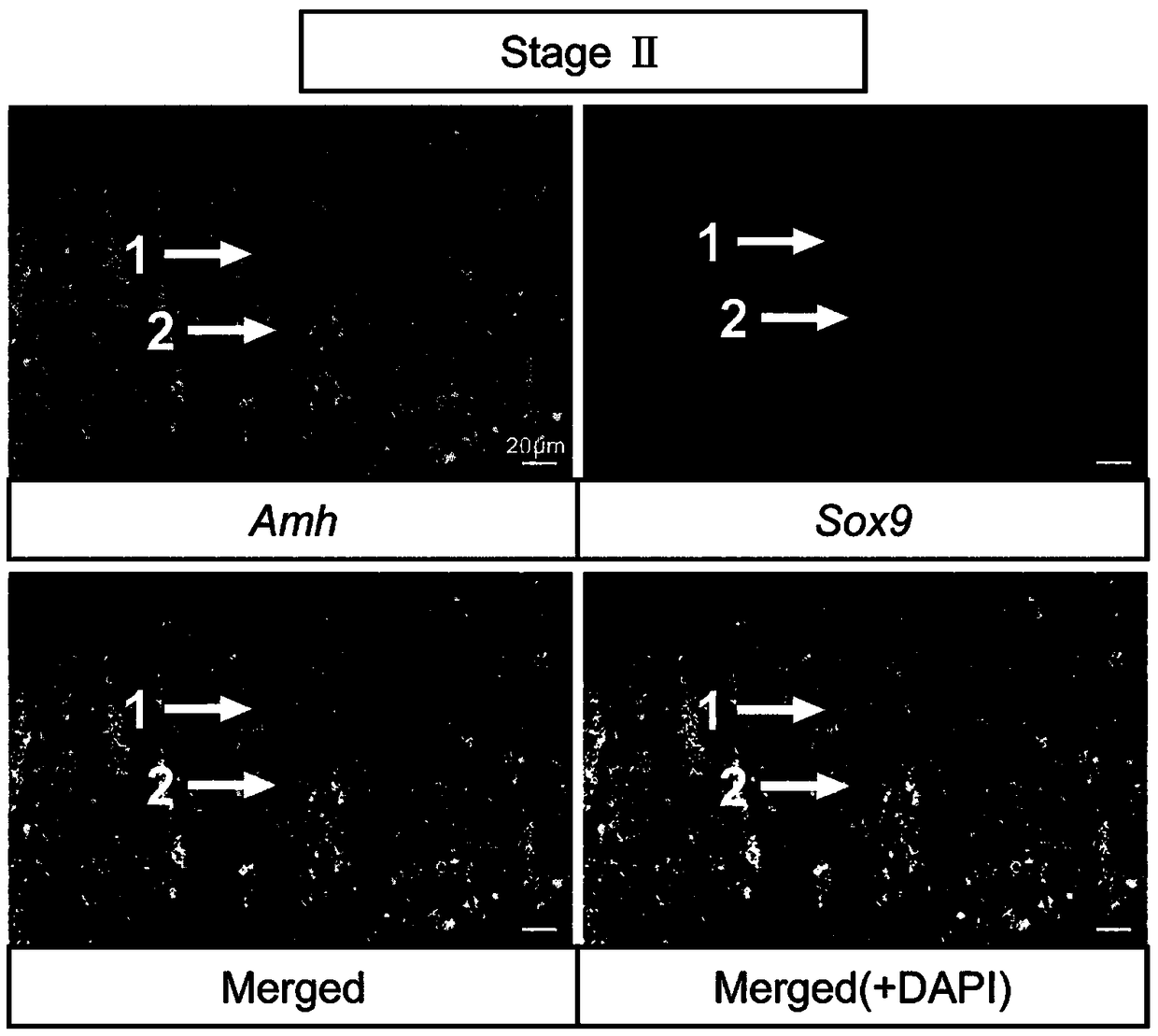
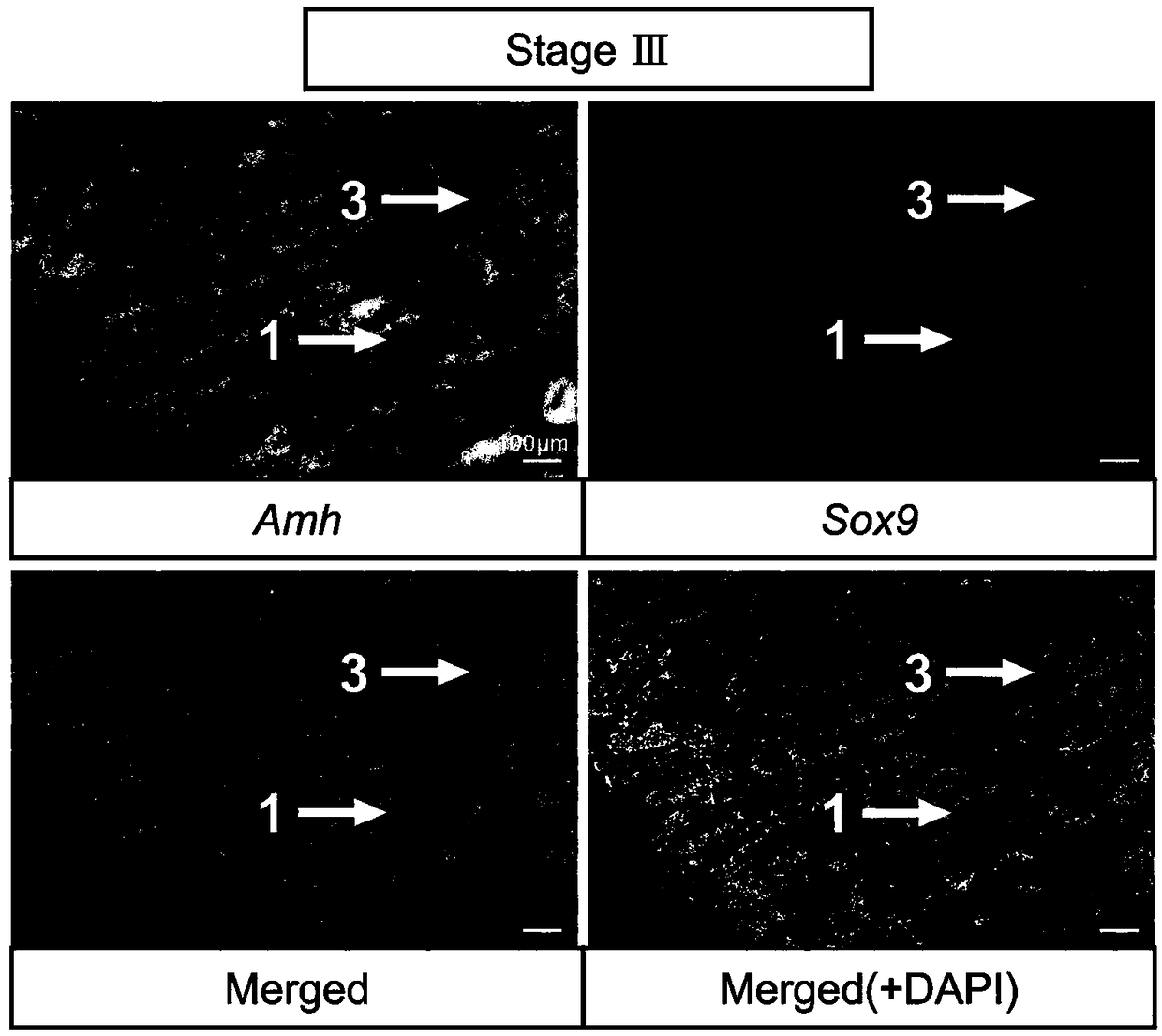
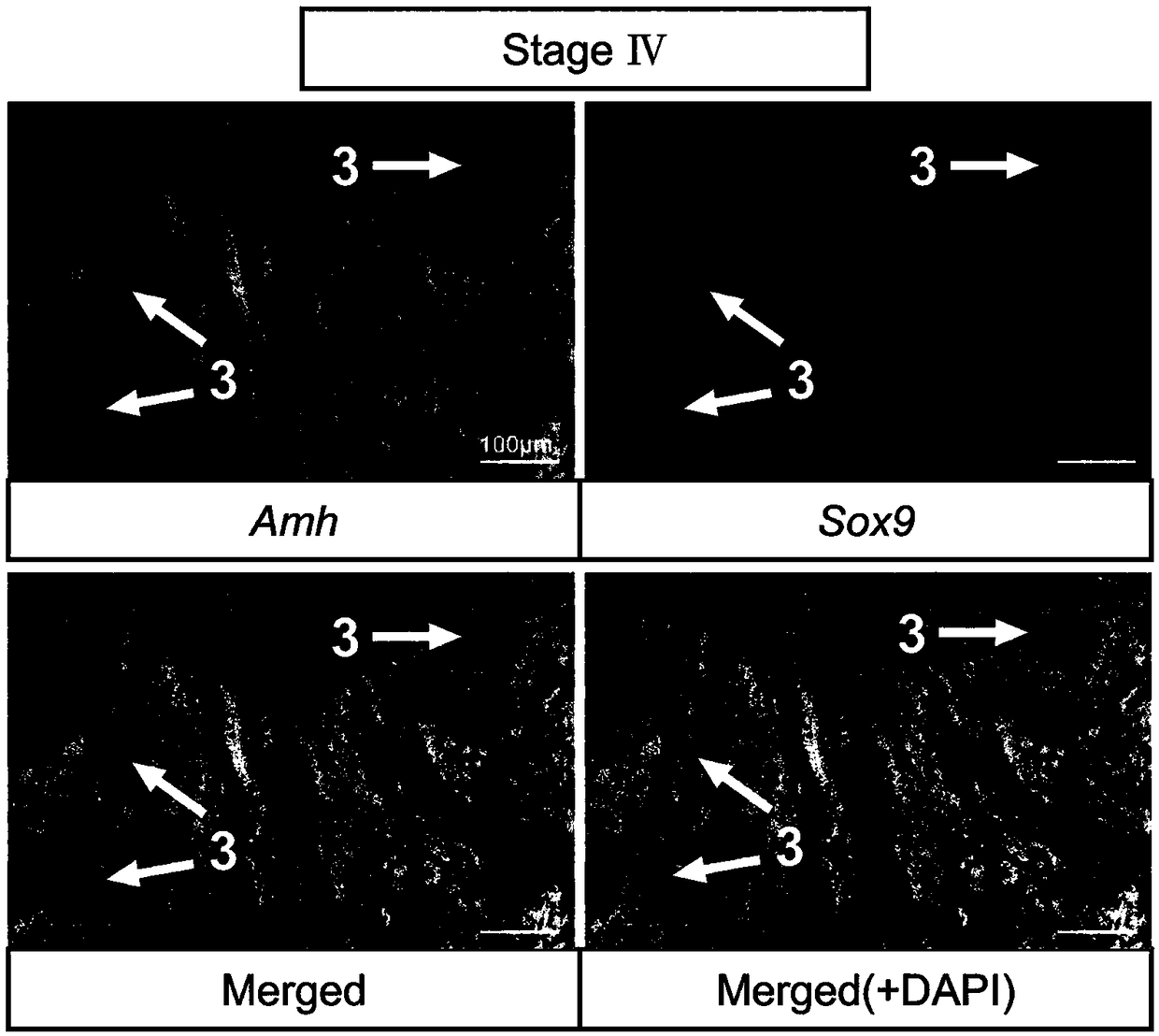
[0046] Methods for labeling germ cells and somatic cells of turbot seminiferous epithelium at different developmental stages include:
[0047] 1) Probe preparation: Amplify Amh / Sox9 / Gsdf fragment from turbot testis cDNA, primer Amh forward sequence: TCCACATTCTCACTCTCGAT, reverse sequence: AGAGTCTCACCATCTCCCTT; Sox9 forward sequence: GACTTTGGAGCCGTGGACAT, reverse sequence: TCACGGTCTGGACAGTTGTG; Gsdf forward sequence: TGAAAGAACCTGCAGCCTCTG, reverse sequence: TTACTCTTTGCTGGGCTGCTG; recovered with 0.98% agarose gel, subcloned and ligated to PGEM-T easy vector, transformed into E. coli competent, selected positive clones and sequenced, picked the correct selection The single clone in the insertion direction, the plasmid is extracted in a medium amount; the plasmids are linearized with restriction endonucleases (NcoI, SpeI), the linearized fragments are purified by the PCR product recovery purification kit and dissolved in DEPC treated water, and the nucleic acid quantifier is used Qua...
Embodiment 2
[0057] Methods for labeling germ cells and somatic cells of turbot seminiferous epithelium at different developmental stages include:
[0058] Probe preparation: Gene in situ hybridization probes used to identify different cell types of turbot seminal epithelium at different developmental stages are shown in Table 1. Amh / Sox9 / Gsdf fragment was amplified from turbot testis cDNA, 1.00 % Agarose electrophoresis gel, subcloned and ligated to PGEM-T easy vector, transformed into E. coli competent, selected positive clones and sequenced, picked a single clone with the correct insertion direction, and extracted a medium amount of plasmid;
[0059] Restriction enzymes (NcoI, SpeI) were used to linearize plasmids, and the linearized fragments were purified by PCR product recovery and purification kit and dissolved in DEPC water, and quantified by nucleic acid quantifier; T7 and SP6 RNA transcriptase were used to synthesize the reverse Sense and sense probes, reaction substances and their co...
Embodiment 3
[0080] The method for labeling germ cells and somatic cells at different developmental stages of turbot seminal epithelium includes: probe preparation, section preparation, and fluorescence in situ hybridization, which specifically includes the following steps:
[0081] Probe preparation: Gene in situ hybridization probes used to identify different cell types of turbot seminal epithelium at different developmental stages are shown in Table 1. Amh / Sox9 / Gsdf fragment was amplified from turbot testis cDNA, 1.05 % Agarose electrophoresis gel, subcloned and ligated to PGEM-T easy vector, transformed into E. coli competent, selected positive clones and sequenced, picked a single clone with the correct insertion direction, and extracted the plasmid in a medium amount; Dicer (NcoI, SpeI) were linearized plasmids, and the linearized fragments were purified by PCR product recovery and purification kit and dissolved in DEPC water, and quantified by nucleic acid quantifier; T7 and SP6 RNA tra...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Thickness | aaaaa | aaaaa |
| Thickness | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com