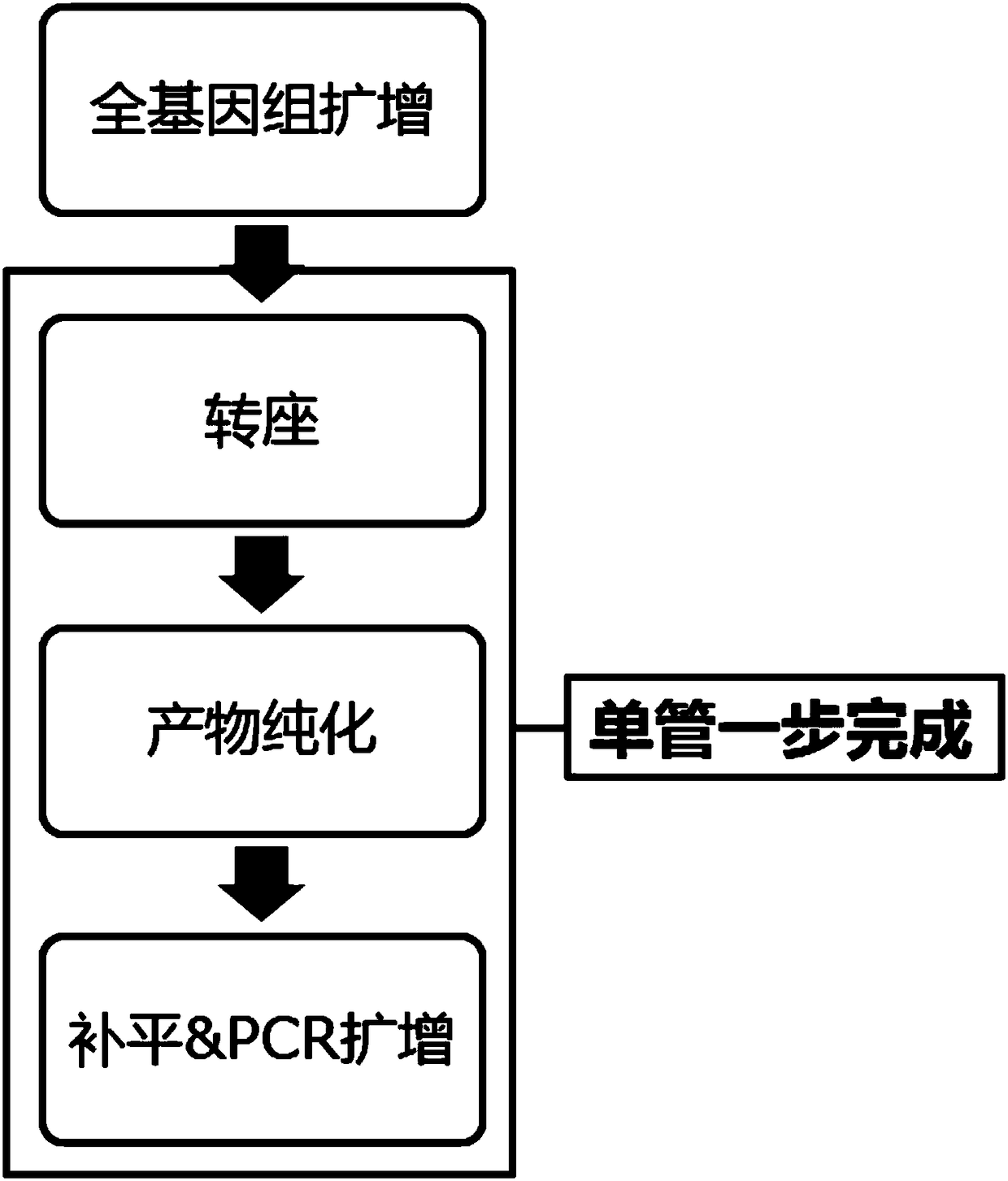
Single-cell genome copy number variation detection method and kit
A detection method and single-cell technology, applied in the biological field, can solve problems such as false positives, false positive results, and different amounts of sequencing data
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0126] 1.1 Single cell picking
[0127] Pick single amniocytes using a mouth pipette or flow cytometer.
[0128] 1.2 Single cell expansion
[0130] 1.2.1.1 According to the number of reactions, mix Cell Lysis Buffer and Cell Lysis Enzyme to prepare the cell lysis mixture.
[0131] Table 1
[0132] Cell Lysis Mixture
volume
Cell Lysis Buffer
2.5μL
Cell Lysis Enzymes
0.05μL
total capacity
2.55 μL
[0133] 1.2.1.2 Collect single cells in a PCR tube containing 2.5 μL of cell lysis mixture.
[0134] 1.2.1.3 Incubate the sample in a preheated PCR machine under the following conditions:
[0135] Table 2
[0136]
[0137] 1.2.1.4 After the procedure ends, briefly centrifuge to collect the reaction solution.
[0138] 1.2.2 Pre-amplification
[0139] 1.2.2.1 Mix Pre-Amp Buffer and Pre-Amp Enzyme Mix to prepare the pre-amplification mixture.
[0140] table 3
[0141] preamplification mix
...
Embodiment 2~4
[0190] 1.1 Single cell picking
[0191] Pick single amniocytes using a mouth pipette or flow cytometer.
[0192] 1.2 Single cell expansion
[0193] 1.2.1 Cell Lysis
[0194] 1.2.1.1 According to the number of reactions, mix Cell Lysis Buffer and Cell Lysis Enzyme to prepare the cell lysis mixture.
[0195] Table 10
[0196] Cell Lysis Mixture
volume
Cell Lysis Buffer
2.5μL
Cell Lysis Enzymes
0.05μL
total capacity
2.55 μL
[0197] 1.2.1.2 Collect single cells in a PCR tube containing 2.5 μL of cell lysis mixture.
[0198] 1.2.1.3 Incubate the sample in a preheated PCR machine under the following conditions:
[0199] Table 11
[0200]
[0201] 1.2.1.4 After the procedure ends, briefly centrifuge to collect the reaction solution.
[0202] 1.2.2 Pre-amplification
[0203] 1.2.2.1 Mix Pre-Amp Buffer and Pre-Amp Enzyme Mix to prepare the pre-amplification mixture.
[0204] Table 12
[0205] preamplification ...
Embodiment 5
[0288] Embodiment 5 data analysis
[0289] Table 19
[0290]
[0291]
[0292]
[0293] Analysis conclusion:
[0294] From the above analysis results, it can be seen that the data quality Q30 and uniformity of the method in this case are significantly higher than that of the comparison method, and a smaller data volume (0.05Gb) and lower coverage (1.2%) are achieved. Chromosomal microamplification and microdeletion detection.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com