Preparation and application of AtAGM2 and AtAGM3 coding gene and enzyme
A coding gene and coding technology, which is applied in application, genetic engineering, plant genetic improvement, etc., can solve the problems of expensive hexose phosphate and cumbersome production steps
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0075] Example 1 Cloning of full-length genes of acetylglucosamine phosphate mutase AtAGM2 and AtAGM3
[0076] After analyzing the hexose phosphate mutase genes of Arabidopsis in The National Center for Biotechnology Information (NCBI) database, two genes in the hexose mutase family whose functions have not been clarified were selected. After sequence analysis, the primers were designed as follows:
[0077] Agm2-F: 5'-GCGTCCATGGAAGGAAAGGTTTTCCAAAAAC-3'; Agm2-R: 5'-ATATATGCGGCCGCGGAGGAGTGAGTG-3' to amplify the gene sequence of Atagm2. Agm3-F: 5'-AACTCCATGGCGTCGACTTCAACATCATC-3'; Agm3-R: 5'-ACTGCTCGAGAGATTGACCTCCGATGTAA-3' to amplify the gene sequence of Atagm3.
[0078] The mRNA of Arabidopsis leaves was extracted according to the operation steps of the RNA extraction kit (Bomad Biotechnology, Cat. No. RN0112). PCR amplification was carried out using the reversed cDNA of the extracted Arabidopsis RNA as a template. The PCR reaction conditions were: 94°C for 2 min, 1 cycle; 9...
Embodiment 2
[0079] Example 2 Gene sequence analysis of acetylglucosamine phosphate mutase AtAGM2 and AtAGM3
[0080] The results of the sequencing were analyzed using the Basic Local Alignment Search Tool (BLAST) in the GenBank database, and the Vector NTI Suite 8.0 software was used for multiple sequence alignment and sequence information analysis.
[0081] The coding region of the obtained acetylglucosamine phosphate mutase gene 2 (named AtAGM2) is 1878 bp long, and its nucleotide sequence is shown in SEQ ID NO 1. AtAGM2 encodes 625 amino acids and a stop codon, its amino acid sequence is shown in SEQ ID NO 3, the theoretical protein molecular weight is 67.0kDa, and the predicted isoelectric point is 6.1. The nucleotide sequence of AtAGM2 is located on chromosome 5 of Arabidopsis thaliana (locus-tag="AT5G17530") in the Arabidopsis genome.
[0082] The coding region of the obtained acetylglucosamine phosphate mutase gene 3 (named AtAGM3) is 1872 bp long, and its nucleotide sequence is s...
Embodiment 3
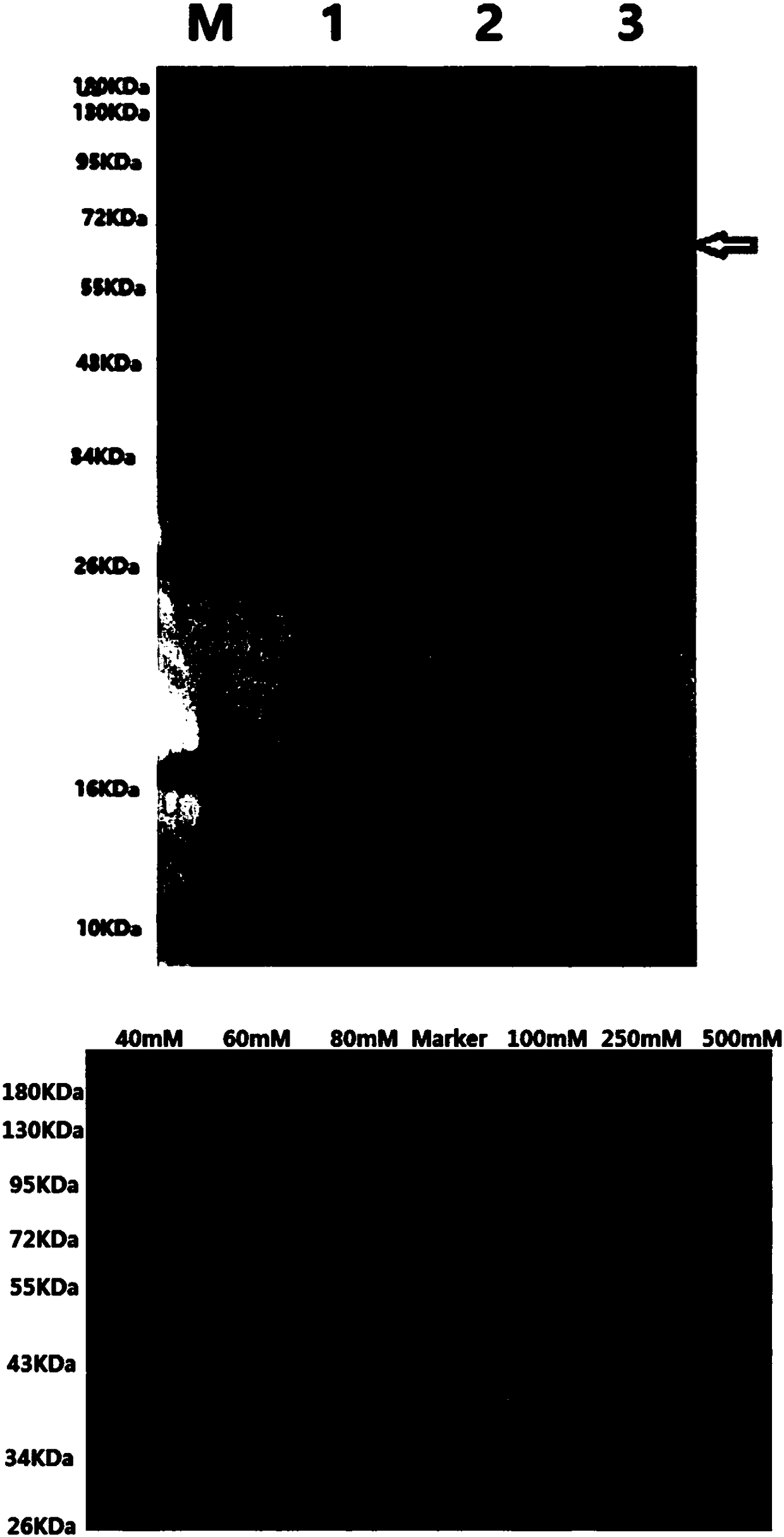
[0086] Example 3 Recombinant expression and purification of AtAGM2 and AtAGM3 genes in Escherichia coli
[0087] Sequencing results showed that the AtAGM2 gene shown in SEQ ID NO 1 was inserted into pET28a, and the insertion direction was correct, which proved that the constructed recombinant plasmid was correct, and the recombinant plasmid was named pET28a-AtAGM2.
[0088] At the same time, the recombinant plasmid of AtAGM3 was also constructed successfully. The AtAGM3 gene shown in SEQ ID NO 3 was inserted into pET28a, and the insertion direction was correct. The recombinant plasmid was named pET28a-AtAGM3.
[0089] Transform pET28a-AtAGM2 (or pET28a-AtAGM3) into Escherichia coli strain BL21(DE3) for induced expression. Add the overnight cultured seed solution to fresh LB medium with 1% inoculum size for expansion at 37°C, add IPTG when the OD600nm of the bacterial solution is 0.6-0.8, make the final concentration 0.5mM, and carry out at 16°C Induce overnight. After the cell...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com