Real-time fluorescent PCR detection method of rhododendron bud wilt and its special set of reagents
A fusarium wilt and rhododendron technology, applied in the biological field, can solve the problems of difficulty in meeting the detection requirements of fast, accurate and sensitive identification, high professional quality requirements of detection personnel, time-consuming and labor-intensive problems, etc., to achieve significant application promotion value and avoid human factors Interference, sensitivity-enhancing effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0061] Embodiment 1, the preparation of the complete set of reagents that detects rhododendron bud wilt
[0062] The complete set of reagents for the detection of Rhododendron wilt bacterium provided by the present embodiment is made up of a primer pair named SA and a probe named SA-P. SA is composed of single-stranded DNA named SA-F and SA-R respectively, and the molar ratio of the two is 1:1. SA can be amplified from the genomic DNA of Rhododendron fusarium wilt to obtain the sequence shown in sequence 4 in the sequence table. DNA fragments. The sequences of each primer are as follows:
[0063] SA-F (forward primer, sequence 1 of the sequence listing): 5'-TGAGATAGCACCCTTTGTTTATGAGT-3';
[0064] SA-R (reverse primer, sequence 2 of the sequence listing): 5'-TGCTACTGCAAAGGGTTTTTCA-3';
[0065] The nucleotide sequence of the probe SAP is shown in Sequence 3 of the Sequence Listing, with a fluorescent reporter group FAM at the 5' end and a fluorescent quencher group MGB at the...
Embodiment 2
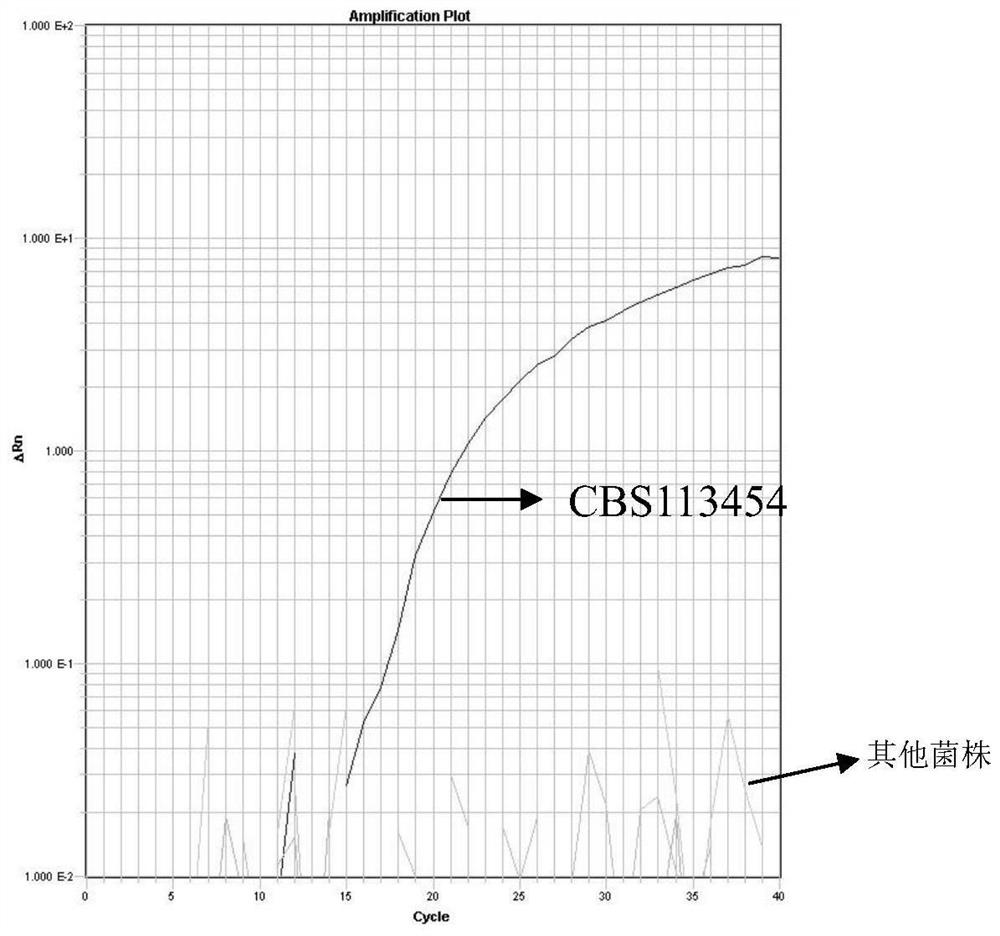
[0068] The specificity of the complete set of reagents of the detection rhododendron bud wilt bacterium of embodiment 2, embodiment 1
[0069] Samples to be tested: Rhododendron bud blight Pycnostysanus azaleae CBS113454, rhododendron wilt CBS 280.53, apple shell color monospore canker Botryosphaeria stevensii MYA 3697, apple canker Cytospora cincta ATCC 28972, soybean southern canker bacterium Diaporthe phaseolorum var.meridional4alis 6 , American-Australian stone fruit brown rot fungus Monilinia fructicola MF, grape stem blight fungus Phoma glomerata ATCC96757, pea foot rot fungus Phoma pinodella ATCC38814, oak sudden death fungus Phytophthora ramorumMYA 3238, soybean Phytophthora sojae ATCC200094, Pythium ATCC3 splendor 、苹果黑星病菌Venturia inaequalisATCC 11096、Verticillium albo-atrum CBS 102464、 Verticillium alfalfae VAA2、Verticillium dahliae VD1、Verticillium nubilum CBS 456.51、Verticilliumtricorpus CBS 447.54,菌除Verticillium alfalfae VAA2、 Verticillium dahliae VD1与美澳型核果褐腐病菌 Moni...
Embodiment 3
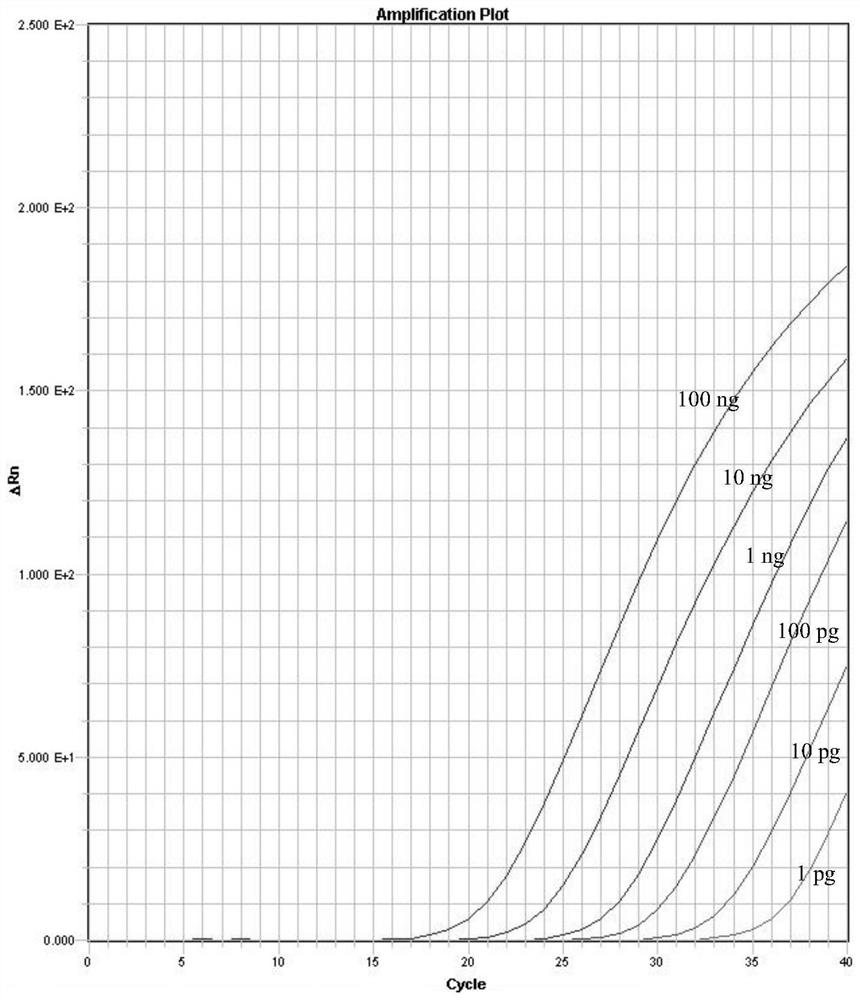
[0075] The sensitivity of the complete set of reagents of the detection rhododendron bud wilt bacterium of embodiment 3, embodiment 1
[0076] 1. Extract the genomic DNA of rhododendron bud blight CBS113454 bacterial strain (Netherlands Microorganism Culture Collection (CBS)), and use the genomic DNA as a template after gradient dilution.
[0077] 2. Using the genomic DNA obtained in step 1 as a template, perform real-time fluorescent PCR.
[0078] Reaction system (10μL): 5μL Universal PCR Master Mix, 0.5 μL SA-F (10 μmol / L), 0.5 μL SA-R (10 μmol / L), 0.5 μL SA-P (10 μmol / L), 1 μL genomic DNA, made up to 10 μL. In this reaction system, the concentration of SA-F is 0.5 μmol / L, the concentration of SA-R is 0.5 μmol / L, and the concentration of SA-P is 0.5 μmol / L. An equal volume of water was used instead of genomic DNA as a blank control.
[0079] There are 1-8 reaction system systems in total, and in reaction system 1, the DNA content of the template is 10ng.
[0080] In re...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com