Kit for detecting human cytomegalovirus
A technology of human cytomegalovirus and cytomegalovirus, which is applied in the determination/inspection of microorganisms, biochemical equipment and methods, etc., can solve the problems of incomparable quantitative results, missed detection of clinical patients, and no traceability, etc., to achieve Suitable for popularization and application, high sensitivity and good specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Example 1 Preparation of a kit for detecting human cytomegalovirus
[0034] The composition of kit of the present invention:
[0035] (1) 20ul reaction solution 1: 50mM tris(hydroxymethyl)methylglycine (pH8.0), 100mM potassium acetate, 0.6uM primer 1, 0.6uM primer 2, 0.3uM primer 3, 0.3uM primer 4, 0.25uM Probe 1, 0.25uM Probe 2, 10U Taq enzyme, 0.5U UNG, 0.3mMdATP, 0.3mMdGTP, 0.3mMdCTP, 0.3mMdUTP, 0.1% sodium azide;
[0036] The sequences of 4 specific primers and 2 specific probes are:
[0037] Primer 1: 5'-AACTGCCTCACCCACCTGC-3' (SEQ ID No.1);
[0038] Primer 2: 5'-AACGATGGAGGACGACCAACAC-3' (SEQ ID No.2);
[0039] Primer 3: 5'-GAAGGCTCATGGCAAGAAAG-3' (SEQ ID No.3);
[0040] Primer 4: 5'-CTCACTCAGTGTGGCAAAGG-3' (SEQ ID No.4);
[0041] Probe 1: FAM- AAACACCATCTTTCCGGAGGTGCGGT-BHQ1 (SEQ ID No. 5);
[0042] Probe 2: ROX-CTTGAGGTTGTCCAGGTGAGCCAG-BHQ2 (SEQ ID No. 6).
[0043] (2) 10ul reaction solution 2: including 1.5mM Mn(OAc) 2 and 0.1% sodium azide.
[0044] (...
Embodiment 2
[0050] Example 2 Human cytomegalovirus nucleic acid extraction and amplification method
[0051] The operation steps of using the kit prepared in Example 1 to detect human giant cell DNA in clinical urine samples are:
[0052] (1) Sample extraction
[0053] 1. Take 1ml of urine and add an equal volume of sample concentrate D, vortex evenly, centrifuge at 12000rpm for 5min, discard the supernatant; add 600µl normal saline, vortex mix, and set aside.
[0054] 2. At the same time, take 600 μl negative quality control, strong positive quality control, weak positive quality control, standard A, B, C, D and the sample to be tested, add 20 μl DNA internal standard respectively, cooperate with Zhengzhou Antu Bioengineering Co., Ltd. magnetic bead method nucleic acid extraction reagent to extract nucleic acid DNA in each sample and set aside.
[0055] (2) Reagent preparation
[0056] According to the number of samples to be tested, negative quality control, strong positive quality c...
Embodiment 3
[0066] Embodiment 3 Sensitivity experiment of the kit of the present invention
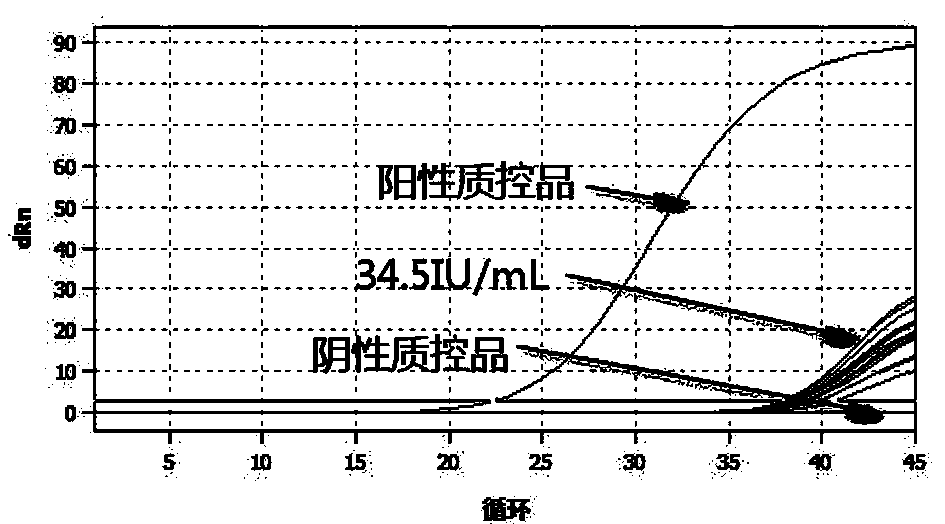
[0067] Use negative urine to dilute HCMV-positive clinical samples (quantity can be traced to WHO) to 34.5IU, use the kit prepared in Example 1 to detect 20 samples, and the obtained amplification curve is as follows figure 1 shown. From figure 1 It can be seen that the kit of the present invention detects 20 positive clinical samples of 34.5IU / mL, and all 20 are detected, which proves that the detection limit of the kit of the present invention can reach 34.5IU / mL, and its sensitivity is very high.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com