Recombinant escherichia coli capable of high-yielding L-methionine and application thereof
A technology for recombining Escherichia coli and methionine, which is applied in the direction of recombinant DNA technology, bacteria, and microbial-based methods, and can solve problems such as the lack of in-depth and detailed research on the fermentation and production of methionine
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
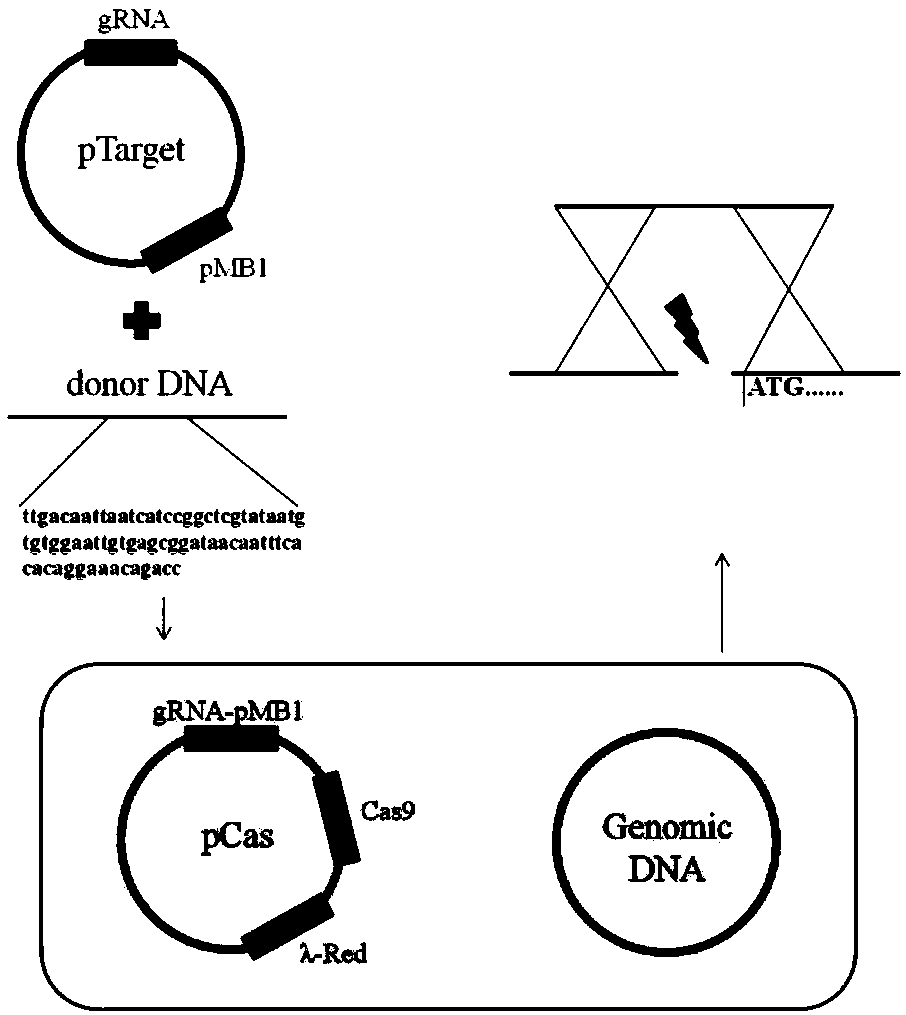
[0046] Example 1: Global search—L-methionine synthesis gene in situ promoter replacement in the genome
[0047] (1) E.coli W3110ΔmetJΔmetI / pTrc99A / metA* / yjeH (Jiangfeng Huang et al.2016.Metabolic engineering of Escherichia coli for microbial production of L-methionine.Biotechnology and Bioengineering.114:843-851) was used as the starting strain, and the global Search strategy, using CRISPR-Cas9-mediated gene editing technology (Yu Jiang et al.2015MultigeneEditing in the Escherichia coli Genome via the CRISPR-Cas9System.AppliedEnvironmental Microbiology.81:2506-2514), using the trc promoter derived from pTrc99A ( The nucleotide sequence is shown in SEQ ID No.1), the original promoters of all genes involved in L-methionine biosynthesis are replaced in the genome, the genes involved and the corresponding pathways are shown in Table 1, identified in Key genes affecting L-methionine in the starting strain.
[0048] Table 1 Genes and corresponding pathways involved in gene editing ...
Embodiment 2
[0063] Example 2: Global Search—L-Methionine Synthetic Gene in Genome Shake Flask Fermentation Test of Strains with In Situ Promoter Replacement
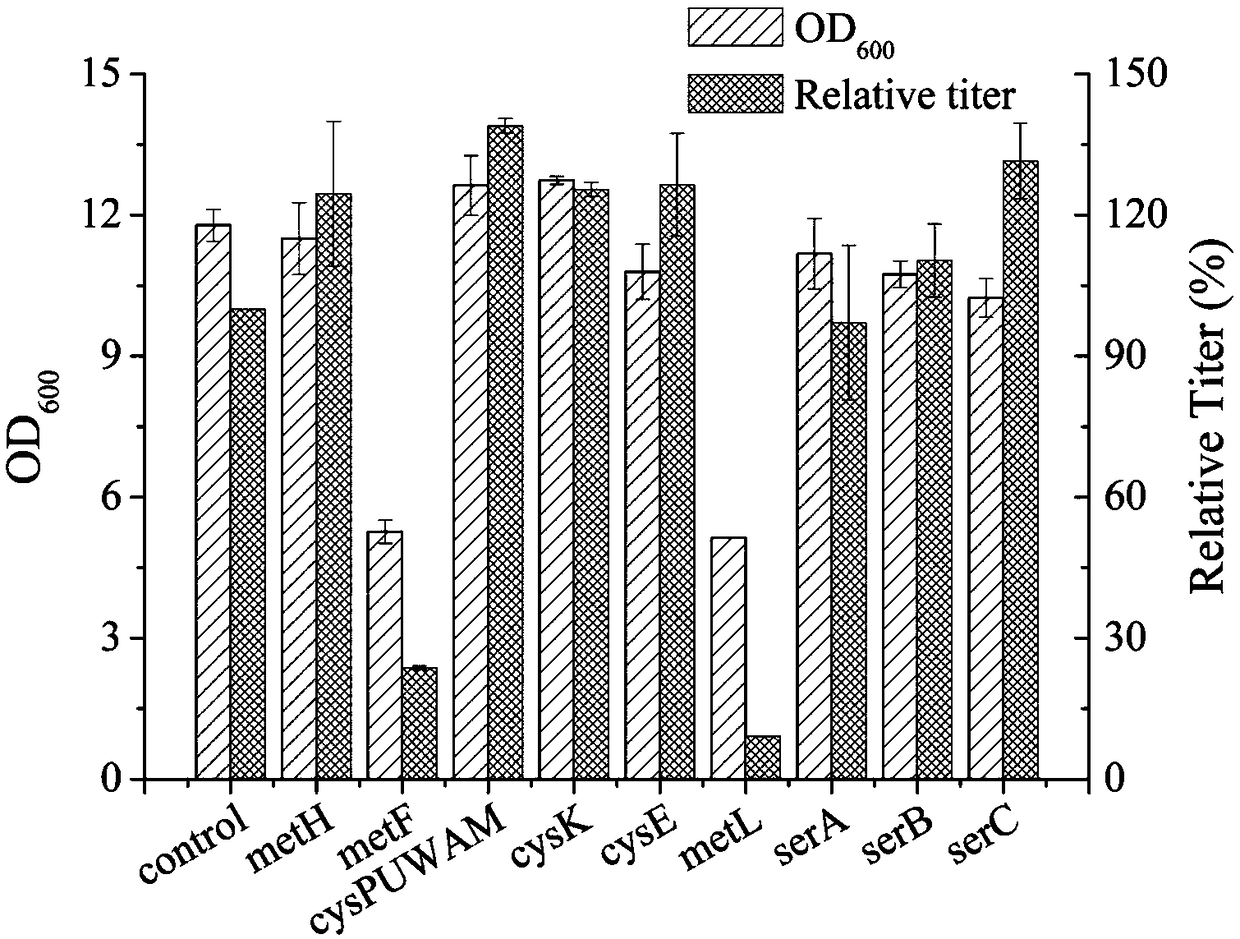
[0064]A series of strains of different genotypes constructed in Example 1 (as shown in Table 4) were inoculated into 10 ml of LB medium containing 50 mg / l of Amp, cultivated at 37° C. and 200 rpm as a preculture, wherein E.coli W3110ΔmetJΔmetI / pTrc99A / metA* / yjeH was a negative control. After 8-12 hours, inoculate 1ml of the preculture into a 500ml shake flask containing 20ml of MS medium containing 50mg / L Amp. Then culture and ferment to OD at 30°C and 150rpm 600 =0.8-1.0, add IPTG with a final concentration of 0.1mM, continue to cultivate for 48h, take 1ml of fermentation broth to measure OD after fermentation 600 Take 1ml of fermentation broth, centrifuge at room temperature at 12000rpm for 3min, dilute the fermentation supernatant 100 times, and analyze the amino acid titer with a fully automatic amino acid analyzer (SYKAM S-4...
Embodiment 3
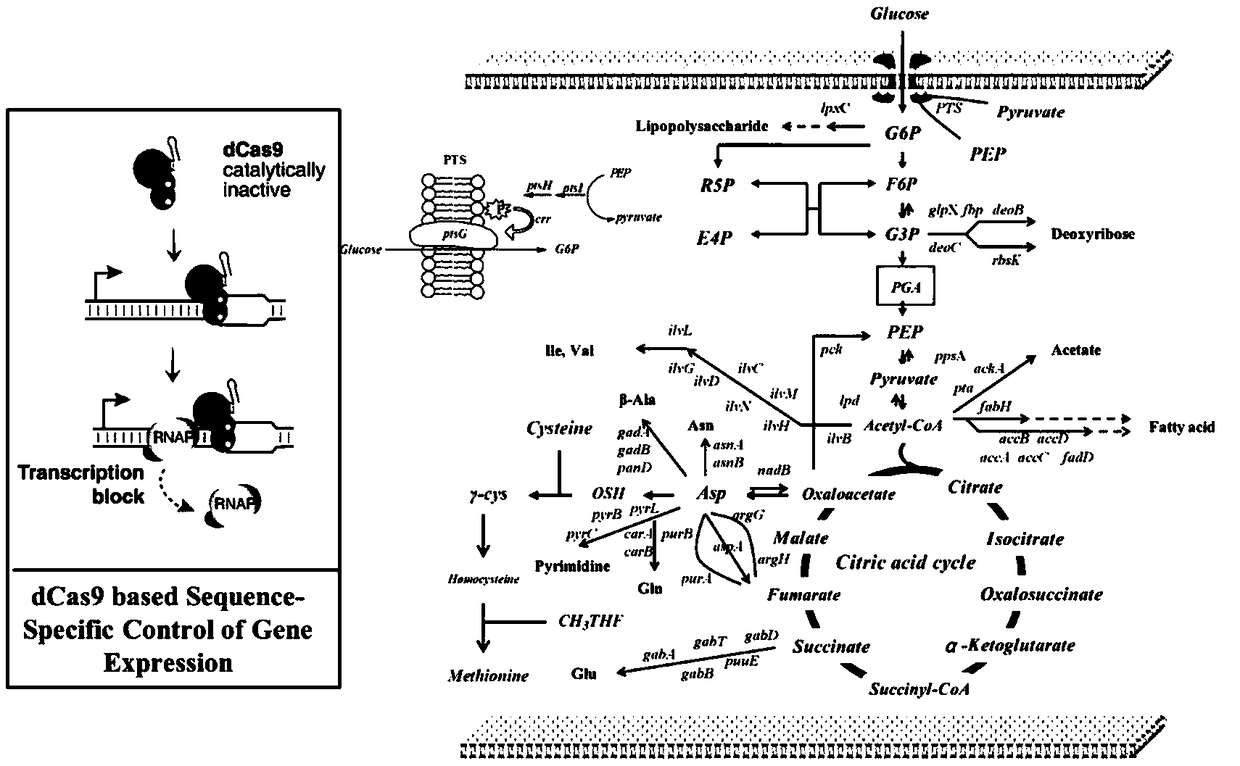
[0067] Example 3: Global Search—Down-regulation of the expression of genes not involved in the synthesis of L-methionine and methionine in the central metabolic pathway
[0068] In order to down-regulate the expression of genes not involved in the biosynthesis of L-methionine and methionine in the central metabolic pathway in the genome, E.coli W3110ΔmetJΔmetI / pTrc99A / metA* / yjeH was used as the starting bacterium to implement CRISPR-dCas9-mediated The sequence-specific gene expression interference technology (CRISPRi) (Qi LS.et al.Repurposed CRISPR as an RNA-Guided Platform for Sequence-Specific Control of GeneExpression.Cell.2013.152(5):1173-1183), this technology can make dCas9 The protein binds to a specific position in the gene coding sequence and reduces gene expression by inhibiting the movement of RNA polymerase. In order to realize this process, the following primers (Table 6) were used to construct pTarget plasmid, pdCas plasmid.
[0069] Use CRISPRi technology to kn...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com