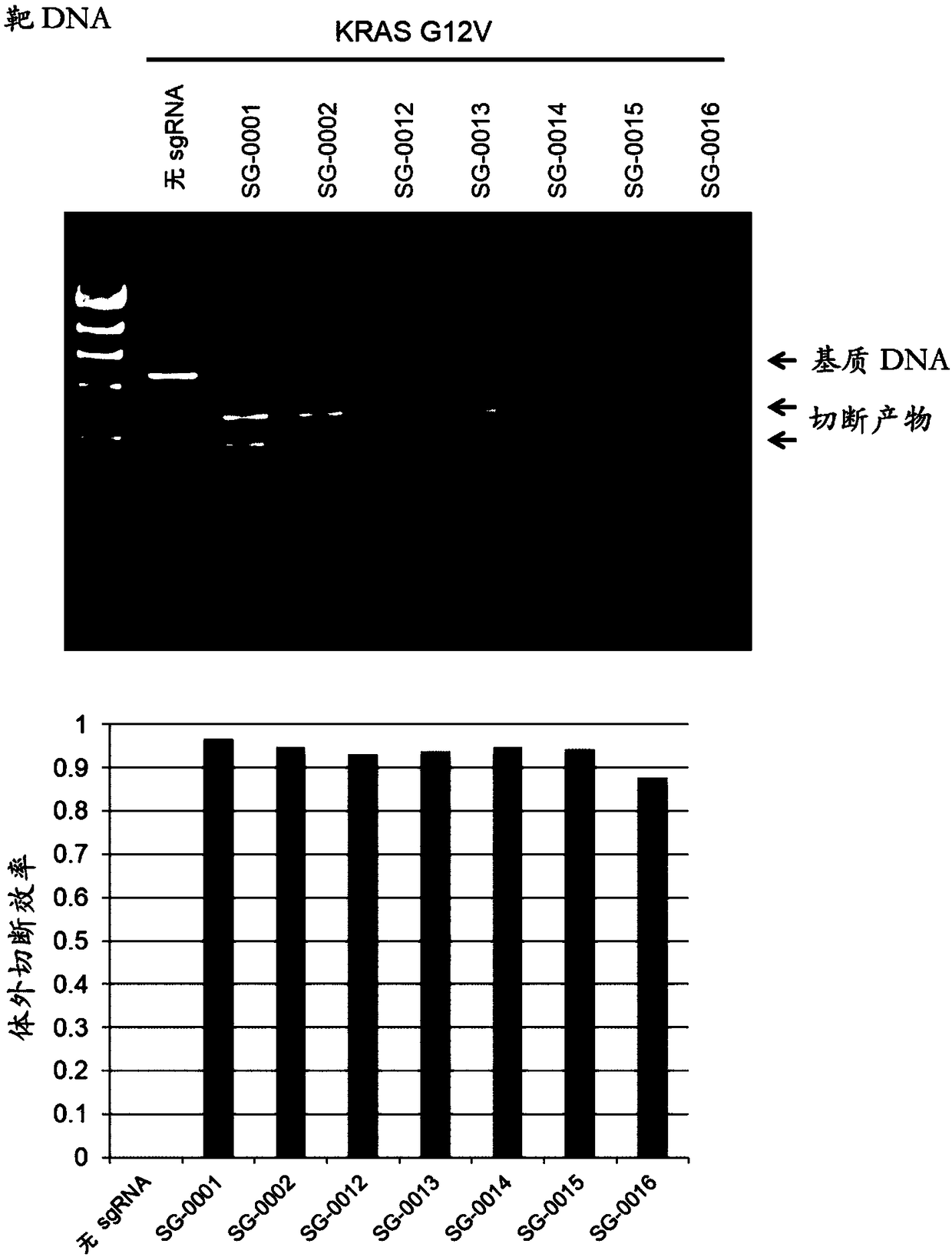
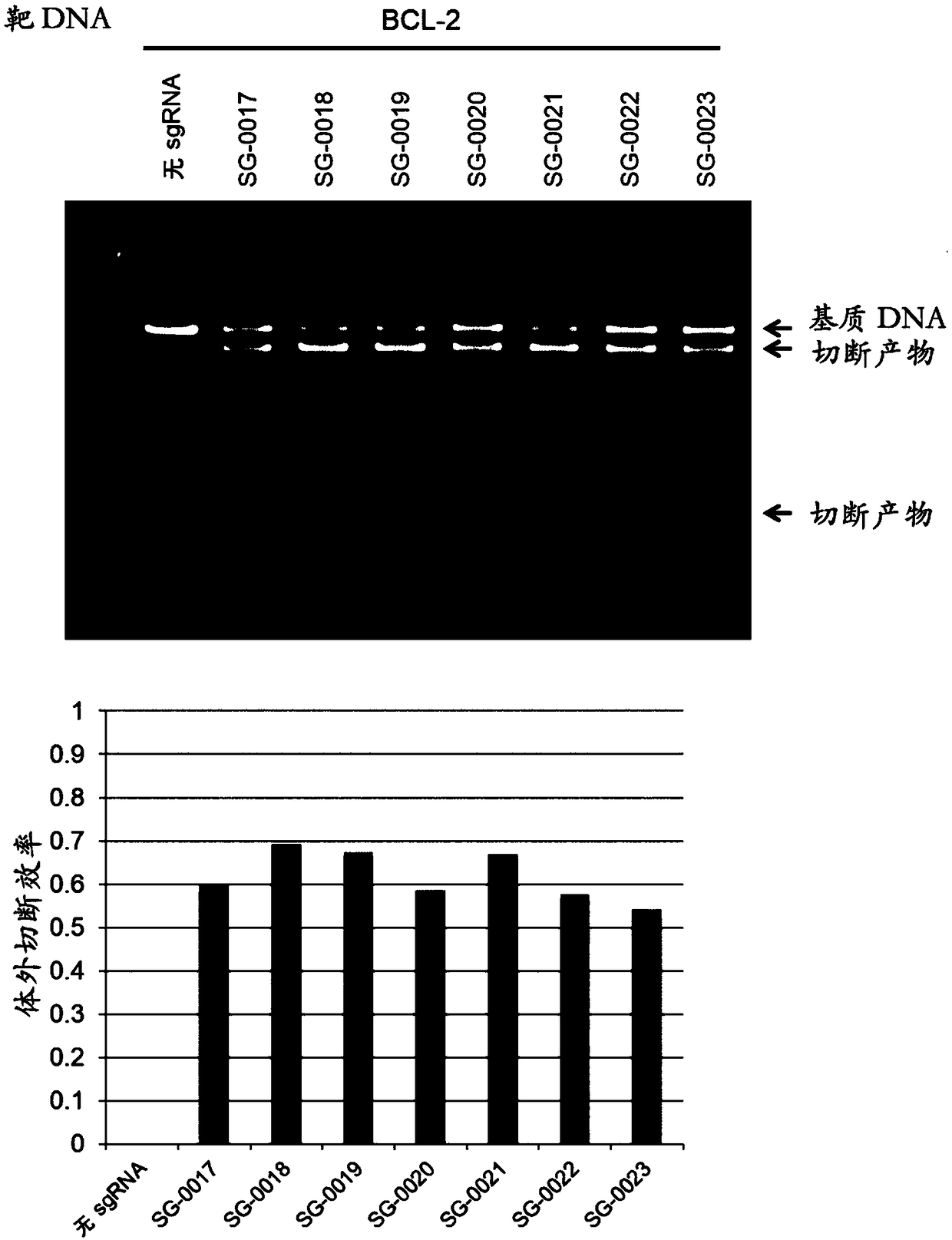
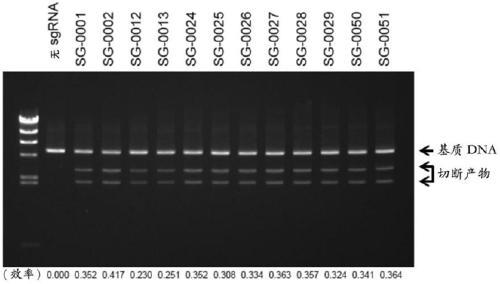
Artificial single guide RNA and use thereof
An independent and atomic technology, applied in the field of artificial single guide RNA, can solve the problems of culture, enzyme reaction time and labor, and natural RNA instability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0351] (Example 1) Synthesis of sgRNA
[0352] The sgRNA shown in Table 1 was synthesized based on the phosphoramidite method and using a nucleic acid synthesizer (The ABI Expedite(R) 8909 Nucleic Acid Synthesis System, Applied Biosystems). The aforementioned synthesis used EMM imide as RNA imide (International Publication No. 2013 / 027843) (the same applies below). The aforementioned imide was deprotected according to a prescribed method, and purified by HPLC. In the following sequence, the underline is the guide region complementary to the target nucleotide sequence, proline diamide imide is introduced into P, lysine diamide imide is introduced into K, and glycylglycine is introduced into X Diamide imide.
[0353] [Table 1]
[0354]
Embodiment 2
[0371] (Embodiment 2) Phenylalanine imide: the synthesis of compound (5)
[0372] Compound (5) was synthesized according to the following scheme.
[0373]
[0374] [[1] Synthesis of compound (1)]
[0375] In Fmoc-phenylalanine (3.0g, 7.7mmol), 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC) (1.78g, 9.3mmol) 1-Hydroxybenzotriazole monohydrate (HOBt) (2.51g, 18.6mmol) in acetonitrile solution (100mL) was added 4-amino-1-butanol (0.83g, 9.3mmol) and stirred overnight at room temperature . After completion of the reaction, the solvent was distilled off under reduced pressure. Dichloromethane was added to the residue, and the residue was washed twice with saturated aqueous sodium bicarbonate solution and once with saturated aqueous sodium chloride solution. The washed organic layer was dried over anhydrous sodium sulfate, and concentrated under reduced pressure to obtain a crude product (4.1 g) of the target compound (1).
[0376] [[2] Synthesis of compou...
Embodiment 3
[0391] (Embodiment 3) Leucine imide: the synthesis of compound (10)
[0392] Compound (10) was synthesized according to the following scheme.
[0393]
[0394] [[1] Synthesis of compound (6)]
[0395] In Fmoc-leucine (3.0g, 8.5mmol), 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC) (1.95g, 10.2mmol), 1-Hydroxybenzotriazole monohydrate (HOBt) (2.75 g, 20.4 mmol) in acetonitrile (90 mL) was added with 4-amino-1-butanol (0.91 g, 10.2 mmol) and stirred overnight at room temperature. After completion of the reaction, the solvent was distilled off under reduced pressure. Dichloromethane was added to the residue, and the residue was washed twice with saturated aqueous sodium bicarbonate solution and once with saturated aqueous sodium chloride solution. The washed organic layer was dried over anhydrous sodium sulfate, and concentrated under reduced pressure to obtain a crude product (4.42 g) of the target compound (6).
[0396] [[2] Synthesis of compound (7)] ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com