Method for detecting leptospira by multiple cross amplification in combination with nano-biosensor
A biosensor and leptospira technology, applied in the field of microbial detection, can solve the problems of complex operation steps and dependence on reagents, and achieve the effect of fast detection speed and excellent detection sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] The feasibility of embodiment 1.MCDA-LFB amplification
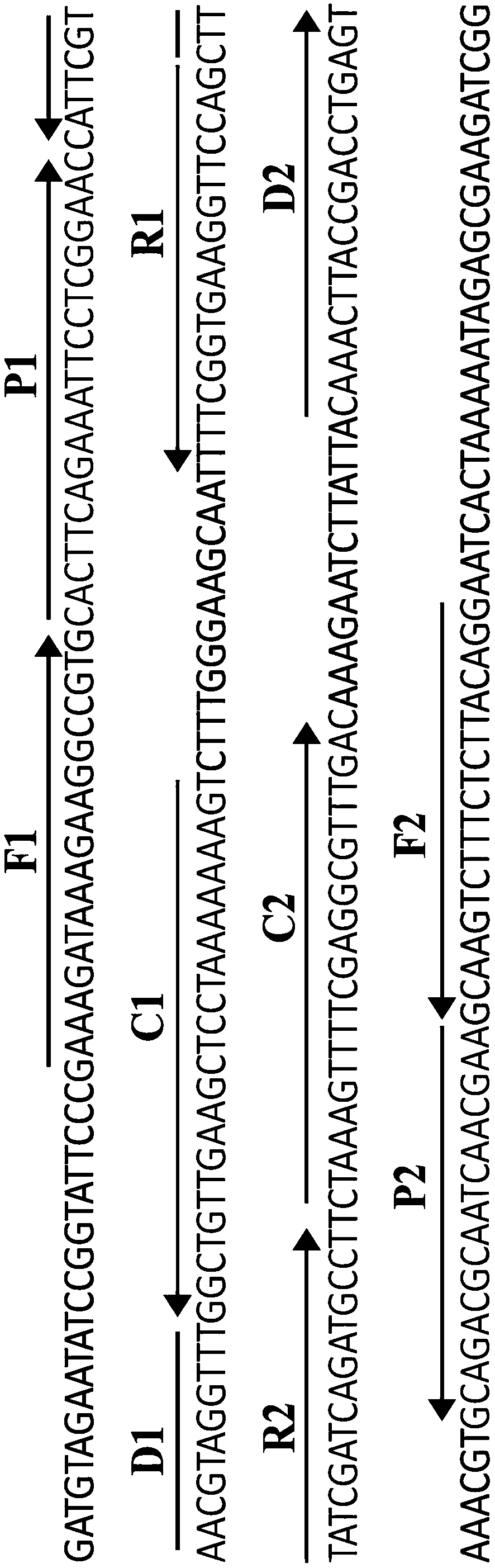
[0049] Standard MCDA reaction system: the concentration of cross primers CP1 and CP2 is 60 pmol, the concentration of displacement primers F1 and F2 is 10 pmol, the concentration of amplification primers R1, R2, D1* and D2 is 30 pmol, and the concentration of amplification primers C1* and C2 The concentration is 20pmol, 10mM Betain, 6mM MgSO 4 , 1 mM dNTP, 12.5 μL of 10×Bst DNA polymerase buffer, 10 U of strand-displacing DNA polymerase, 1 μL of template, and add deionized water to 25 μl. The entire reaction was thermostated at 64°C for 35 minutes.
[0050] After MCDA amplification, two detection methods are used to discriminate MCDA amplification. First, a visible dye (such as MG reagent, leuco malachite green visual reagent) is added to the reaction mixture, and the color of the positive reaction tube changes from colorless to It is bright green, and the negative reaction tube remains colorless. Secondly, MCD...
Embodiment 2
[0053] Embodiment 2. measure the optimal reaction temperature of MCDA technology
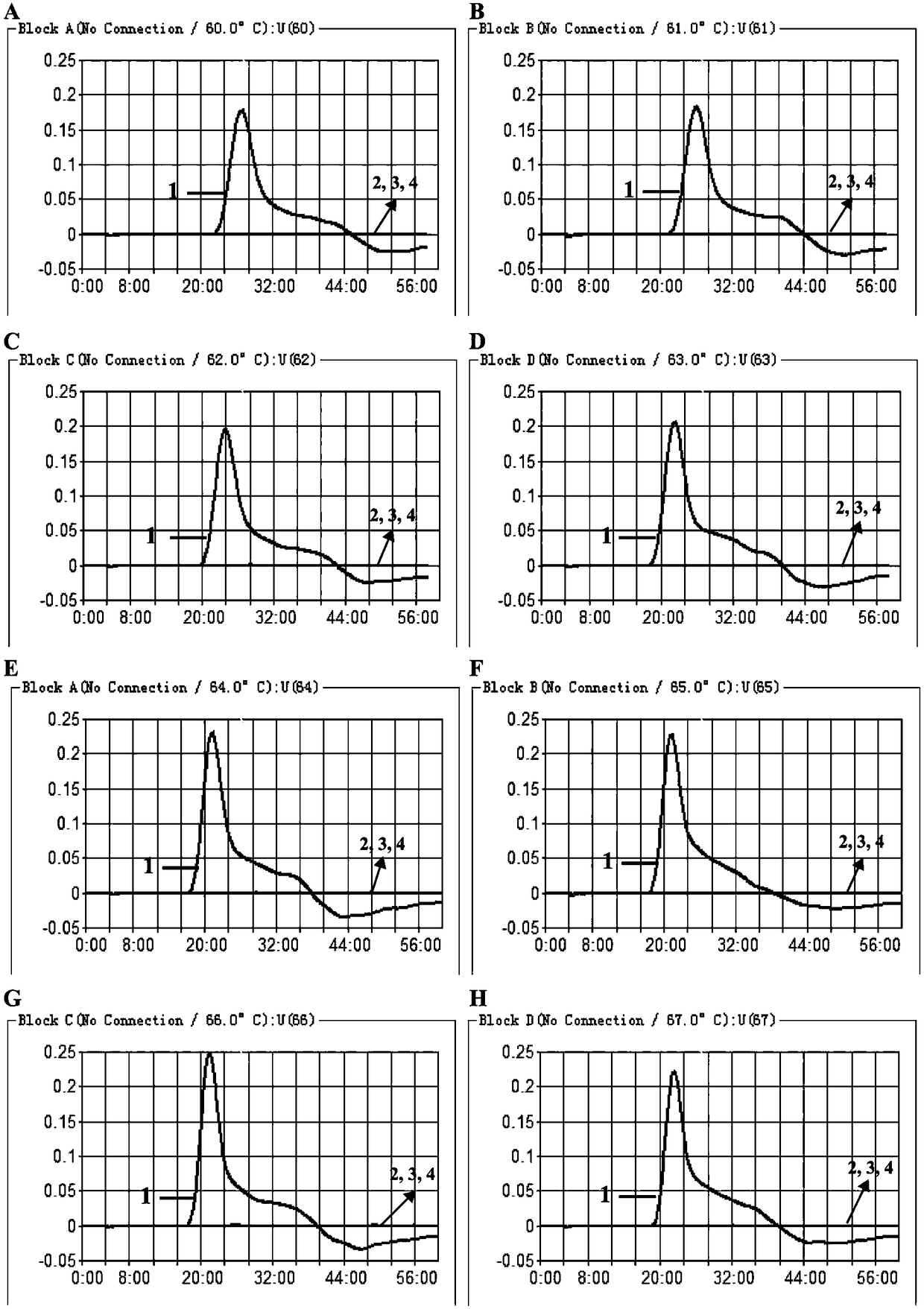
[0054] Under standard reaction system conditions, the DNA template for pathogenic Leptospira and the corresponding MCDA primer designed were added, and the template concentration was 10 pg / ul. The reaction was carried out under constant temperature conditions (60-67°C), and the results were detected by a real-time turbidimeter, and different dynamic curves were obtained at different temperatures, see image 3 . 61-64°C ( image 3 Middle B-E) are recommended as the optimal reaction temperature for MCDA primers. In the follow-up verification of the present invention, 62° C. was selected as the constant temperature condition for MCDA amplification. image 3 It represents the temperature dynamic curve of MCDA primers designed for the lipL41 gene sequence to detect pathogenic Leptospira.
Embodiment 3
[0055] Embodiment 3.MCDA-LFB detects the sensitivity of single target
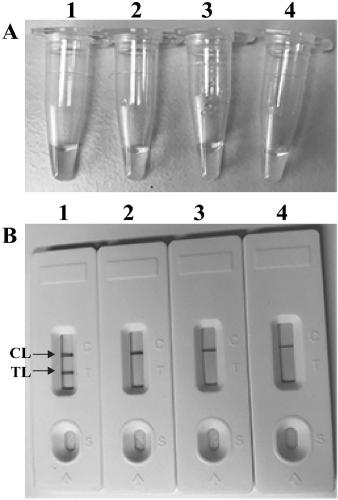
[0056] After using the serially diluted pathogenic Leptospira genomic DNA to carry out the standard MCDA amplification reaction, the LFB detection showed that the detection range of MCDA-LFB was 10 ng-10 fg, and LFB appeared red lines in the TL and CL regions ( Figure 4 Middle A). When the amount of genomic template in the reaction system is reduced to below 10fg, LFB only appears a red line in the CL area, indicating a negative result ( Figure 4 LFB6-8 in A). Figure 4 Middle A is the use of LFB to visually read the MCDA amplification results; Figure 4 LFB 1 to 8 in A respectively represent that the template amount of pathogenic Leptospira is 1x10 5 genomic equivalents(GEs) / reaction; 1x10 4 GEs / reaction; 1x10 3 GEs / reaction; 1x102 GEs / reaction; 1x10 1 GEs / reaction; 1x10 0 GEs / reaction; 1x10 -1 GEs / reaction; 1 µl double distilled water.
[0057] In order to further verify the sensitivity of MCD...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com