Method for rapidly and quantitatively detecting lactobacillus rhamnosus phages
A technology of Lactobacillus rhamnosus and quantitative detection method, which is applied in the field of microorganisms, can solve the problems of no corresponding method for phage detection, high detection cost, and long detection cycle, so as to quickly determine the source and route of phage contamination, and is easy to popularize and use , Improve the effect of timeliness
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Example 1 Preparation of immunomagnetic beads and capture of live cells of Lactobacillus rhamnosus
[0031] The preparation steps of immunomagnetic beads are as follows: 2 mg (200 μL) carboxy-modified magnetic nanobeads (MNB) were placed in a 1.5 mL centrifuge tube, washed twice with 500 μL MEST buffer, and then suspended in 500 μL EDC-NHSS solution. After activation at 37°C for 30 min, MNB was collected by magnetic separation, suspended with 500 μL PBST, washed 3 times, and added to a new sterile 1.5 mL centrifuge tube. Add the specific polyclonal antibody (PcAb-SpaA) (PcAb-SpaA) (100 μL) of Lactobacillus rhamnosus pilus subunit to the solution, incubate at 37°C for 3 h, and discard the supernatant. The PcAb-SpaA-coupled MNB (PcAb-SpaA-MNB) was blocked in PBST containing 1% BSA at 37°C for 45 min, washed 3 times with PBST, and washed with 0.02% NaN 3 Resuspend with 0.5% BSA in PBST (200 μL) to a final concentration of 10 mg / ml, and store at 4°C for later use.
[0032...
Embodiment 2
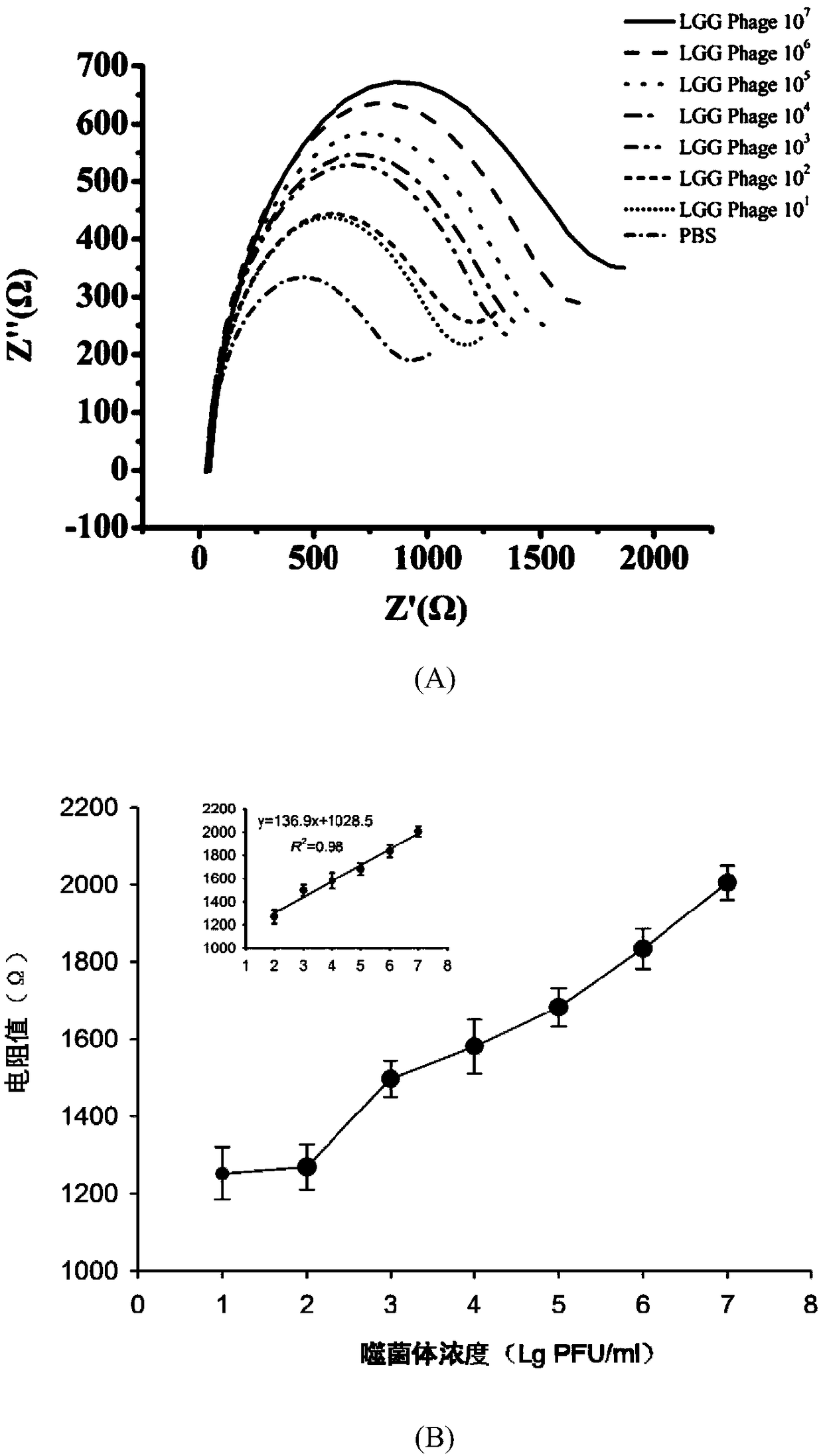
[0035] Example 2 Phage Capture and Electrochemical Signal Detection
[0036] Phage capture Take 100 μL of 109 PFU / mL Lactobacillus rhamnosus (LGG strain) phage diluted to 10 with PBS 7 PFU / mL, 10 6 PFU / mL, 10 5 PFU / mL, 10 4 PFU / mL, 10 3 PFU / mL, 10 2 PFU / mL to prepare phage gradient samples, and do 3 parallels for each concentration; 9 PFU / mL Enterobacter sakazakii phage, diluted to 10 with PBS 7 PFU / mL, 10 6 PFU / mL, 10 5 PFU / mL, 10 4 PFU / mL, 10 3 PFU / mL, 10 2 PFU / mL, three parallels were done for each concentration; PBS without phage was used as a blank control. Take 100 μL of live cell suspension of Lactobacillus rhamnosus and add it to 900 μL of the above-mentioned phage gradient dilution sample, mix well, and let stand at room temperature for 15 minutes for adsorption; take 10 μL of immunomagnetic beads prepared according to (1) and add PcAb-SpaA-MNB Add to each sample, mix well, incubate at 37°C for 60 min; discard the supernatant after magnetic fi...
Embodiment 3
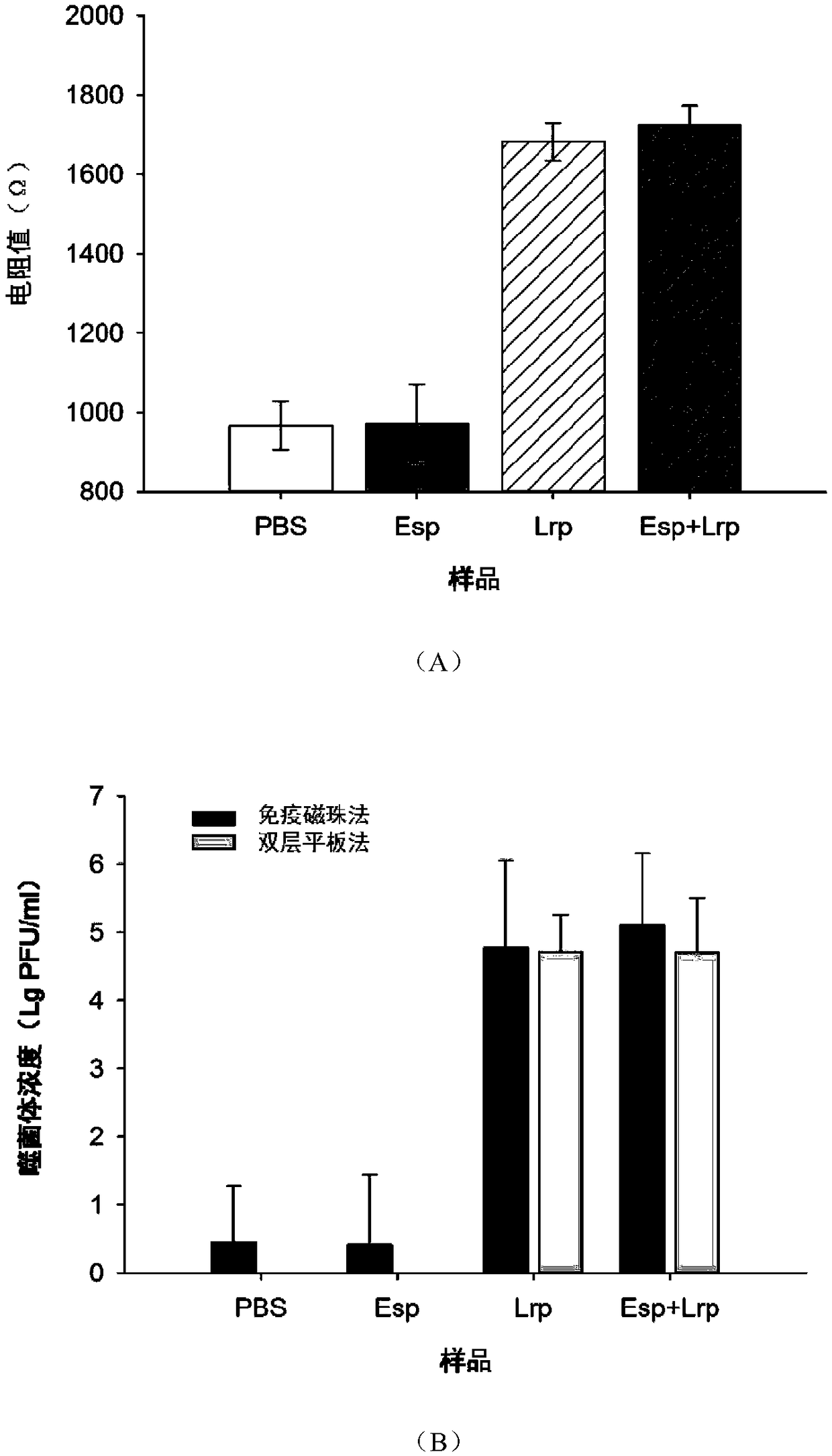
[0039] Embodiment 3 simulated sample detection result
[0040] Take 10μL concentration of 10 7 PFU / mL of Lactobacillus rhamnosus phage and 10 μL of 10 7 Mix PFU / mL Enterobacter sakazakii phage, add 1ml MRS medium and mix thoroughly to make a sample, use MRS without Lactobacillus rhamnosus phage as a negative control; use the traditional double-layer plate method to determine the phage content, specific steps: Take 10 μL of the sample with sterile SM buffer for serial dilution, take 100 μL of the appropriate dilution and mix it with 100 μL of Lactobacillus rhamnosus cultivated overnight at room temperature for 15 min, add 5 mL of MCM semi-solid medium, Quickly pour it onto the bottom plate, wait for the medium to fully solidify, incubate at a constant temperature of 37°C until clear phage plaques appear, and count the number of phage plaques.
[0041] Take 100 μL of live cell suspension of Lactobacillus rhamnosus and add 900 μL of the above sample, mix well, let stand at ro...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com