Method for detecting different beta-casein variants in milk
A detection method and casein technology are applied in the fields of animal breeding and food detection, which can solve problems such as false positive results, and achieve the effects of simple operation, reasonable design and high detection accuracy.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
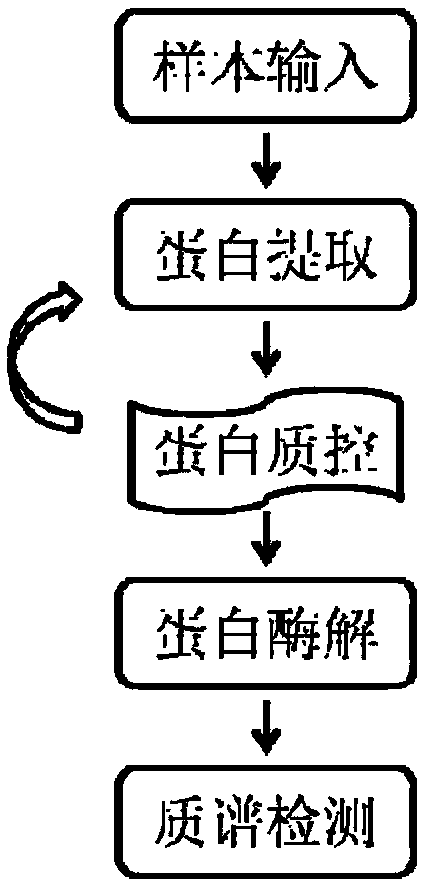
[0072] Example 1: Construction of a method for detecting different β-casein variants
[0073] 1. Protein extraction and quality control of milk or milk powder samples.
[0074] 8M urea was used to fully dissolve the protein in the sample, the protein and oil in the sample were separated by centrifugation, and the obtained protein sample was subjected to dithiothreitol (DTT) reduction and iodoacetamide (IAM) alkylation protection. Coomassie brilliant blue staining (bradford method) was then used to quantify the protein, and sodium dodecylsulfonate-polyacrylamide gel electrophoresis (SDS-PAGE) was used to identify whether the protein band was clear without degradation.
[0075] 2. Carry out enzymatic digestion to the above-mentioned extracted protein;
[0076] Get appropriate amount of above-mentioned sample, utilize the method for ultracentrifugation, the buffer solution in the sample is changed into 25mM ammonium bicarbonate (NH 4 HCO 3 ), in order to facilitate subsequent ...
Embodiment 2
[0081] Embodiment 2: The accuracy and repeatability detection of the method.
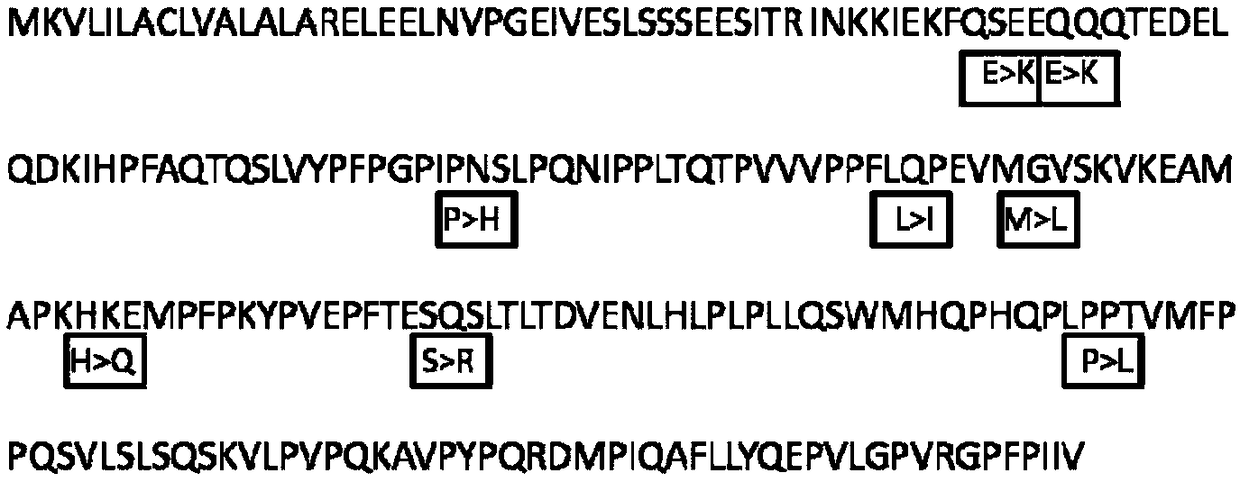
[0082] The dairy cows whose β-casein genotypes have been identified through direct DNA sequencing are used as detection objects, and the method of Example 1 is used to identify different variants of β-casein in milk, and the accuracy rate reaches 100%. It shows that the detection method is accurate and effective.
[0083] Taking milk samples with known β-casein variants as the detection object, this detection method was used for repeated tests, and the results of multiple repeated tests were consistent, indicating that the method has good stability.
Embodiment 3
[0084] Example 3: Detection of actual samples.
[0085] The method of Example 1 of the present invention was used to detect two raw milk samples.
[0086] 1. Protein extraction and quality control;
[0087] (1) Take 50 μl of raw milk sample and add 500 μl of lysate (8M urea, 30mM 4-hydroxyethylpiperazineethanesulfonic acid (Hepes)). Lyse on ice for 10 min. Centrifuge at 4°C, 20000g, 30min, take the supernatant, avoid absorbing oil.
[0088] (2) Add dithiothreitol (DTT) to a final concentration of 10 mM. 56°C water bath for 1h. After taking it out, add iodoacetamide (IAM) quickly to a final concentration of 55mM, and let it stand in a dark room for 1h.
[0089] (3) Proteins were quantified using the Coomassie blue staining method (Bradford method).
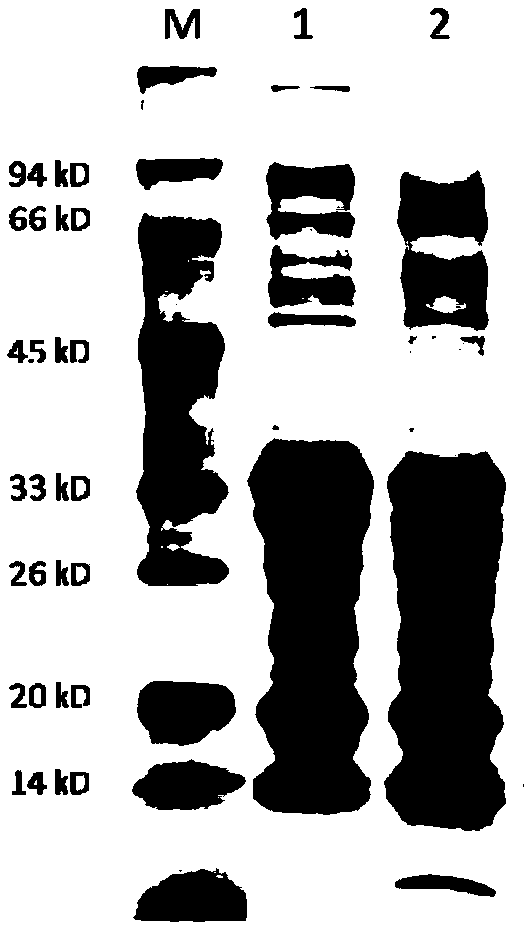
[0090] (4) Carry out sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) to the protein after quantification ( figure 2 ).
[0091] 2. Carry out proteolytic digestion of the above-mentioned extracted prote...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com