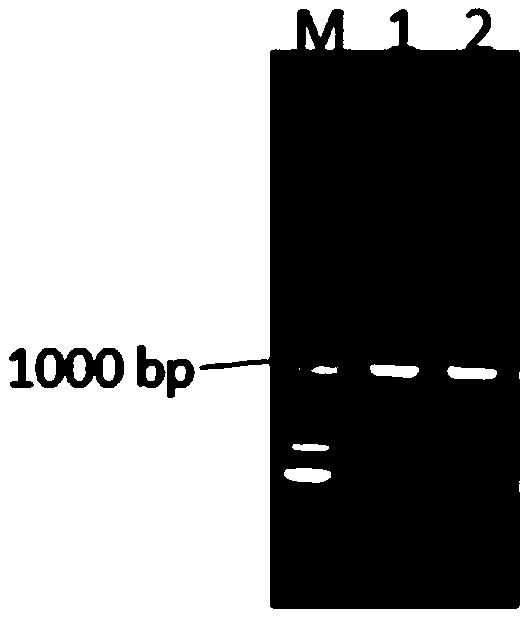
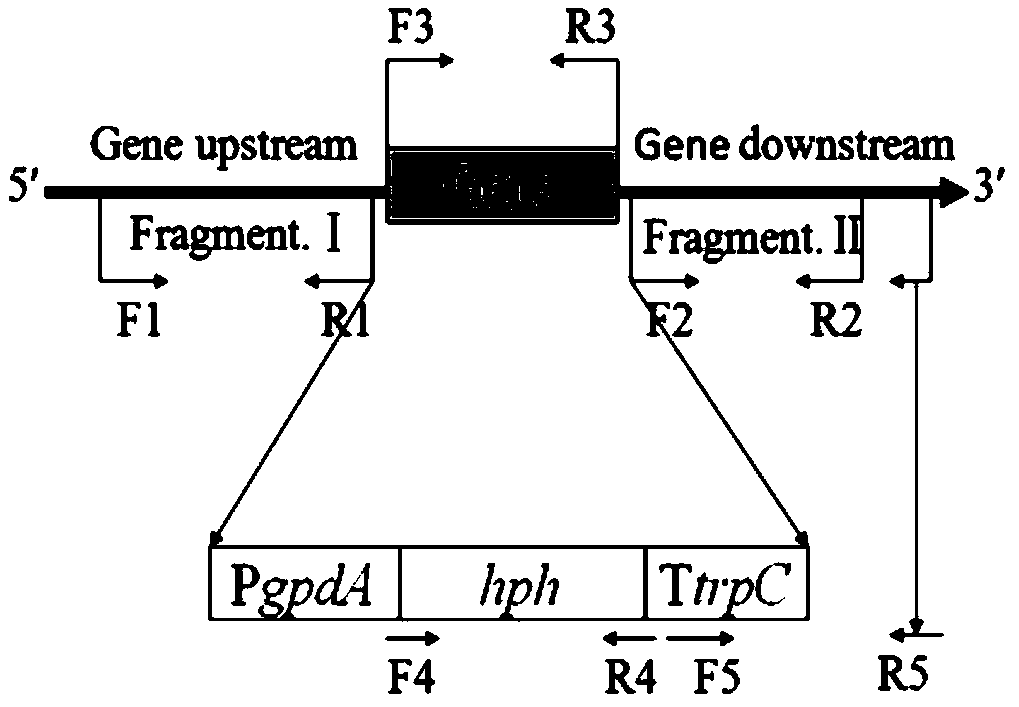
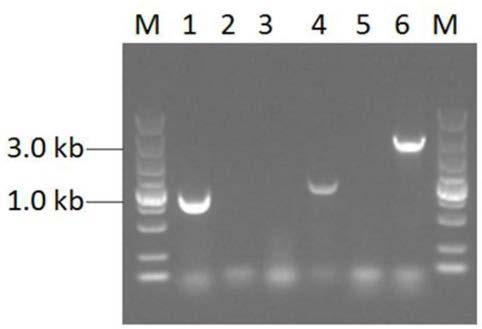
Construction method of high yield monascorubin strain
A technology of high-yield strains and construction methods, applied in the direction of using vectors to introduce foreign genetic material, recombinant DNA technology, etc., can solve the problem of low yield of monascus pigment
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] 1. Construction of binary plasmid knockout vector pHph0380.
[0042] The original vector was the commercial plant binary plasmid pCambia0380. Design a pair of oligonucleotide sequences F&R, which were synthesized by Shanghai Sangon Bioengineering Co., Ltd. This sequence in turn contains the following restriction endonuclease sites (Hind III, Kpn I, Sac I, Pac I, Pme I, Xho I, Xba I, Bgl II). The binary plasmid vector pCambia0380 was cut with restriction endonucleases Hind III and Bgl II. The oligonucleotide sequence was ligated with the restriction vector pCambia0380 by T4 DNA ligase to obtain the binary plasmid vector pCambia0380G.
[0043] Using the plasmid pMD19-PgpdA-hph-TtrpC preserved in the laboratory as a template, the hph expression cassette was amplified with primers PgpdA-Sac I-F&TtrpC-Xho I-R. The hph expression cassette fragment and the binary plasmid vector pCambia0380G were simultaneously digested with restriction endonucleases Sac I and Xho I. The hp...
Embodiment 2
[0096] 1. Construction of binary plasmid knockout vector pHph0380
[0097] The original vector was the commercial plant binary plasmid pCambia0380. Design 1 pair of oligonucleotide sequences F&R, containing in order the following restriction endonuclease sites (Hind III, Kpn I, Sac I, Pac I, Pme I, Xho I, XbaI, Bgl II). The binary plasmid vector pCambia0380 was cut with restriction endonucleases Hind III and Bgl II. The oligonucleotide sequence was connected to the vector by T4 DNA ligase to obtain the binary plasmid vector pCambia0380G.
[0098] Using the plasmid pMD19-PgpdA-hph-TtrpC preserved in the laboratory as a template, the hph expression cassette fragment was amplified with primers PgpdA-Sac I-F&TtrpC-Xho I-R. The promoter fragment and the binary plasmid vector pCambia0380G were digested simultaneously with restriction endonucleases Sac I and Xho I. The oligonucleotide sequence was connected to the vector by T4 DNA ligase to obtain the binary plasmid knockout vecto...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com