ApcE2 protein mutant and application thereof
A protein mutant and mutant technology, applied in the application, bacterial peptides, biochemical equipment and methods, etc., can solve the problems of affecting labeling, deletion modification, insufficient stability, etc., to achieve good solubility, improved solubility, and high unit yield. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
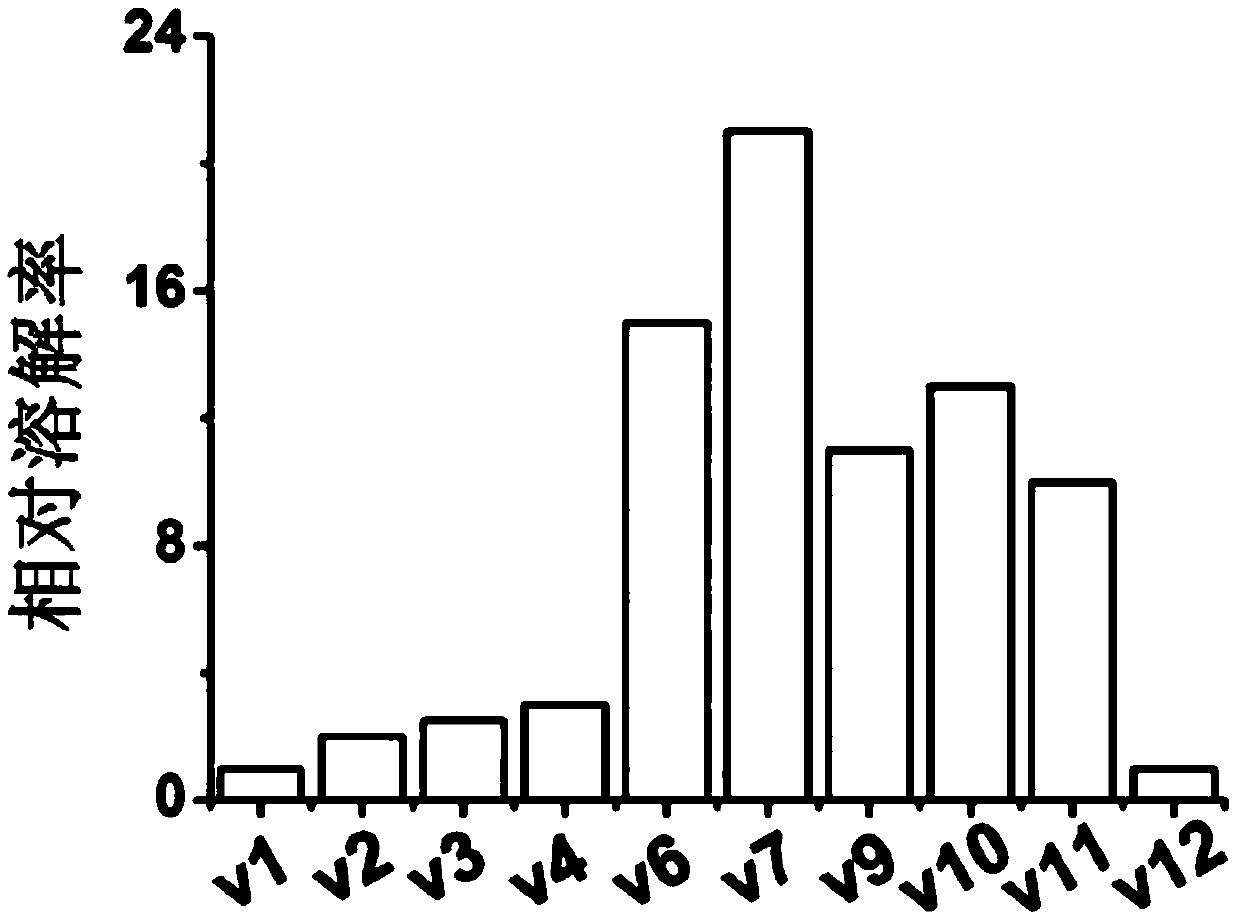
[0046] On the basis of the v0 sequence (amino acid sequence such as SEQ ID No. 1), the hydrophobic amino acid sites (R164K, S165A, L47E, L51N, L54T) outside the loop were mutated to obtain the v1-v4 mutant sequences, amino acid sequences respectively. As shown in SEQ ID No.2 to SEQ ID No.5.
[0047] On the basis of the mutant v4, the hydrophobic amino acid sites (L91T, V92S, L134E, L137D) in the loop were mutated to obtain the v5-v6 mutant sequences respectively, and the amino acid sequences are as shown in SEQ ID No.6~SEQ ID No. 7 is shown.
[0048] On the basis of mutant v6, the hydrophobic amino acid sites (W133Y, K135R) in the loop were mutated to obtain the v7-v8 mutant sequence. The amino acid sequence is shown in SEQ ID No. 8 to SEQ ID No. 9, wherein The v7 mutant was named BDFP3.
[0049] On the basis of mutant v7, the mutant loop structure sequence (Δ88-106, Δ88-114, Δ88-120, Δ88-130) was partially deleted to obtain the v9-v12 mutant sequence. The amino acid sequenc...
Embodiment 2
[0062] Preparation of Phytochrome PΦB and Its Application in Labeling Mammalian Cells
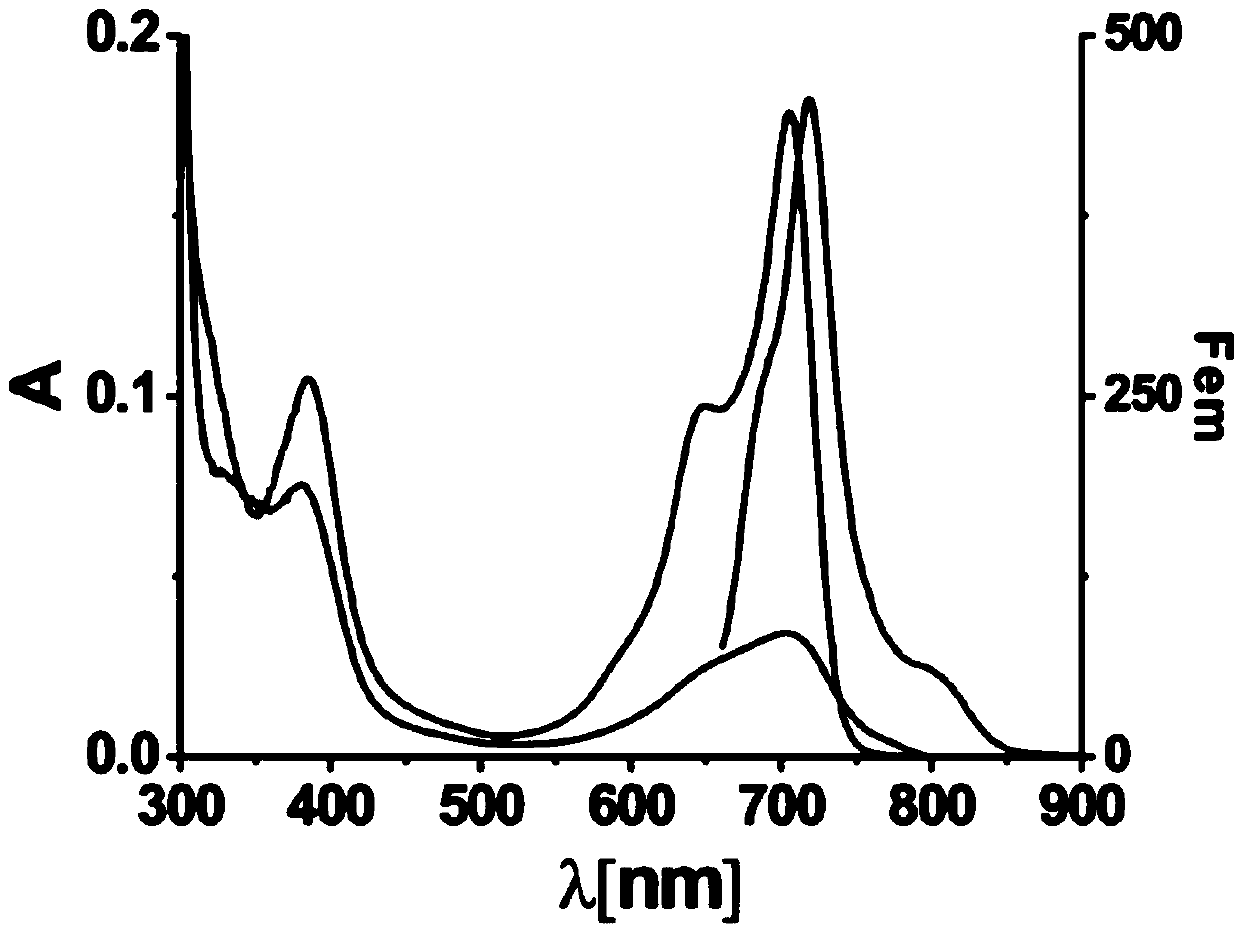
[0063] The gene sequence encoding the mutant was substituted for ApcE2(24-245), co-transformed into Escherichia coli with the heme oxidoreductase gene ho1 and the phytochrome synthase gene hy2, and recombinantly expressed to generate the pigment protein PΦB-mutant, which was extracted and purified. The phytochrome PΦB can be extracted from the chromoprotein PΦB-mutant under denaturing conditions. Pigment crystals can also be obtained by further rotary evaporation.
[0064] Specifically, the purified pigment protein was taken and its absorption characteristic peak was diluted to OD=2, proteinase K with a final concentration of 20 μg / mL was added, and the protein was digested in a water bath at 37° C. for 1 h. The degree of enzymatic hydrolysis was measured by an absorption spectrometer. When the enzymatic hydrolysis was relatively complete, proteinase K was removed to obtain a solution cont...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com