Nucleotide sequence for detecting BRAF gene V600E mutation and application thereof
A nucleic acid sequence and nucleotide sequence technology, which is applied in the nucleotide sequence field of BRAFV600E gene mutation detection, can solve the problems of small difference in Tm value and non-specific amplification, and achieves good effect, high degree of automation and suitable for clinical practice. The effect of application and promotion
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
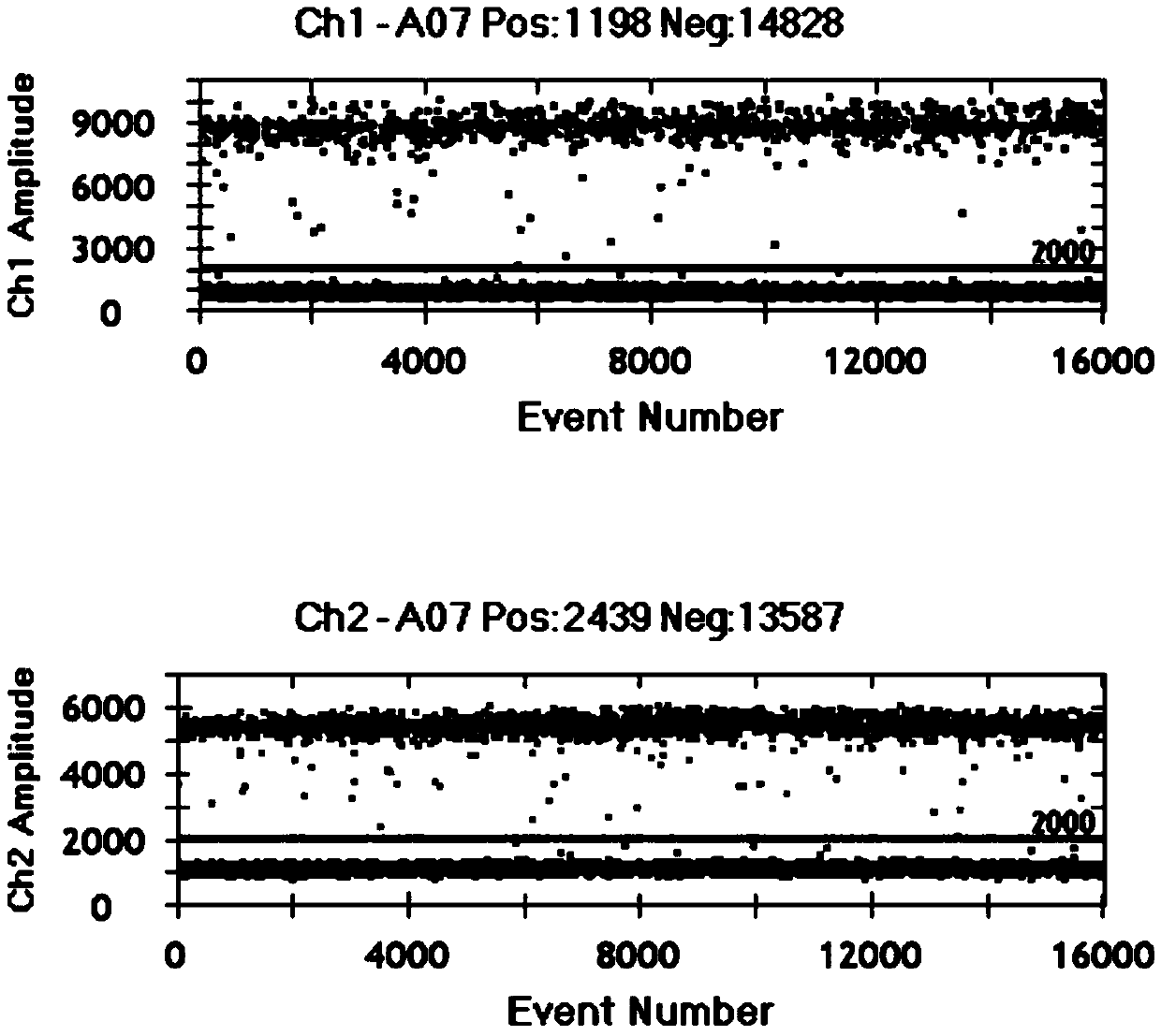
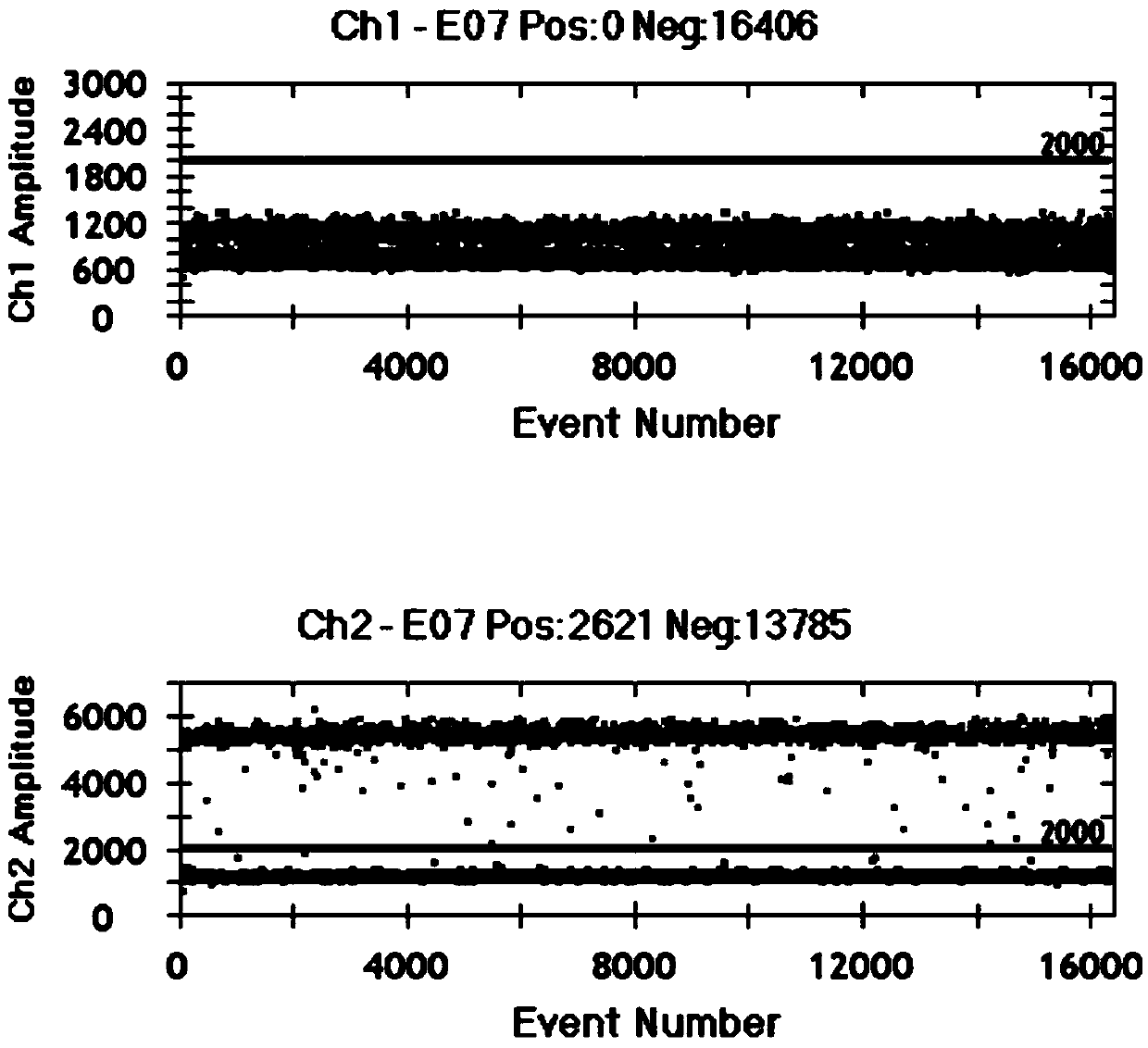
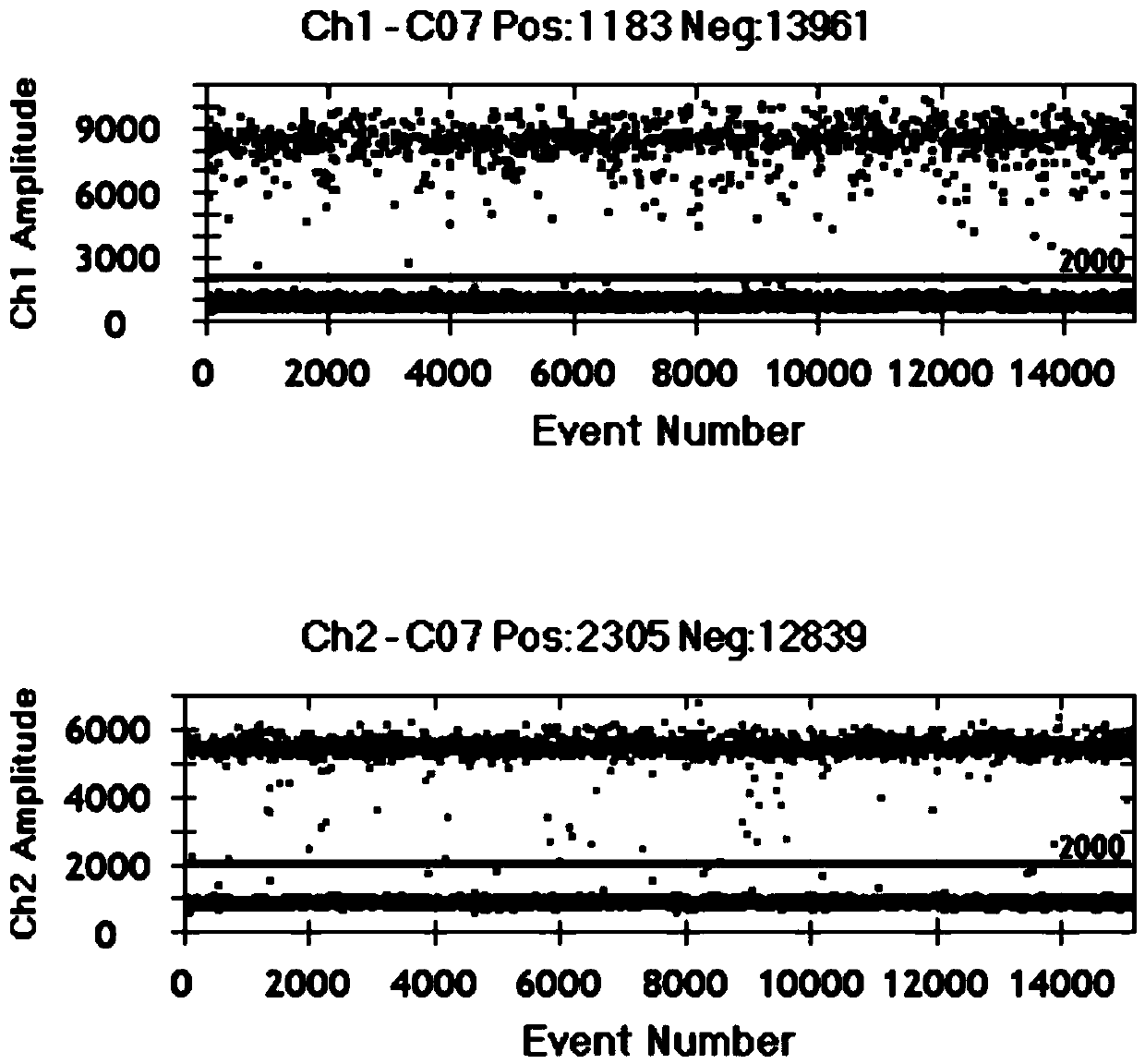
Image
Examples
Embodiment 1
[0037] Example 1: The primers and probes for detecting BRAF were synthesized by Yingwei Jieji (Shanghai) Trading Co., Ltd., and the sequence is as follows:
[0038] Primers for amplifying BRAF:
[0039] F1 (SEQ ID NO: 1): 5'-catgaagacctcacagtaaaa-3';
[0040] R1 (SEQ ID NO: 2): 5'-cagacaactgttcaaactga-3';
[0041] Probes to detect BRAF wild-type:
[0042] P BRAFV600-WT (SEQ ID NO: 3): 5'-cta+ca+g+t+ga+aat+ct-3';
[0043] Among them, the 5' end of the BRAF wild-type probe P BRAFV600-WT (SEQ ID NO: 3) sequence is labeled with a FAM fluorescent group, and the 3' end is labeled with a BHQ1 quencher group. In addition, the 4th, 6th, and The 7th, 8th, 10th, and 13th nucleotides are modified by LNA (indicated by + sign).
[0044] Probes for detecting BRAF mutant genes:
[0045] P V600E-Mut (SEQ ID NO: 4): 5'-cta+ca+g+a+ga+aat+ct-3';
[0046] Among them, the 5' end of the BRAF mutant gene probe P V600E-Mut (SEQ ID NO: 6) is marked with a VIC fluorescent group, and the 3' end is ...
Embodiment 2
[0047] Example 2: Preparation method of BRAF V600E gene mutation detection kit.
[0048] (1) PCR reaction solution: 2*supermix (purchased from Bio-rad Company in the United States, item number 186-3026), which is a 2*PCR reaction premix, which contains DNA polymerase, Mg 2+ , PCR reaction buffer, dATP, dCTP, dTTP and dGTP and other components, stored at -20°C;
[0049] (2) Primer-probe premix solution: Dissolve the primers and probes shown in SEQ ID NO: 1 to 4 in double distilled water, the concentration of each primer is 100 μM, and the concentration of each probe is 10 μM; SEQ ID NO: The mother solutions of ID NO: 1 to 3 nucleic acid sequences were mixed at a volume ratio of 4.5:4.5:25; the mother solutions of SEQ ID NO: 1, SEQ ID NO: 2, and SEQ ID NO: 4 were mixed at a volume ratio of 4.5:4.5: The volume ratio ratio of 25 was mixed; the final primer / probe concentration in the reaction system was 900 / 250nM. Store at -20°C;
[0050] (3) Positive control: a solution contain...
Embodiment 3
[0052] Embodiment 3: detection method.
[0053] Instruments: Eppendorf Mastercycler pro S qualitative PCR instrument, Bio-rad QX200 droplet digital PCR system (including droplet generator, sealing device, droplet reader), BECKMAN 22R desktop micro-refrigerated centrifuge, WH-866 vortex oscillator (Taicang Hualida), low-speed plate centrifuge (Anhui Zhongjia).
[0054] 1. Preparation of BRAF V600E sample, positive control, and negative control DNA template: use QIAamp Circulating Nucleic Acid Kit from QIAGEN (Cat. No. 55114), and operate according to the kit instructions.
[0055] 2. Use 1 pair of specific primers and 2 specific probes to detect the BRAF mutation gene, specifically including the following steps;
[0056] (1) Preparation of PCR reaction solution: Take out the components of the kit from the -20°C refrigerator, thaw at room temperature, and put them on an ice box for later use. Within 10 minutes before adding the sample, configure the PCR reaction solution X ul...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com