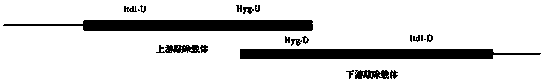Ustilago esculenta teliospore formation related gene Itd1 and application thereof
A kind of technology of wild black powder fungus and teliospore, which is applied to the related gene Itd1 of black powder fungus teliospore formation and its application field, and can solve the unpublished and other problems.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0017] Embodiment 1: wild rice smut ( Ustilago esculenta ) Itd1 gene cloning
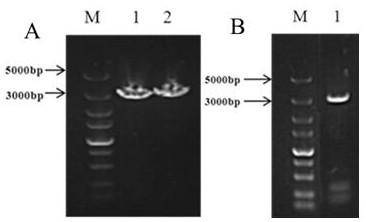
[0018] 1. Cloning of the Itd1 gene: Through the analysis of the transcriptome data of normal Zizania and Glycyrrhizae, combined with the third-generation sequencing data of the Ustilago smut genome, it was predicted that the Ustilago smut gene locus related to teliospore formation was g1449. Using the ORF Finder function of NCBI, the open reading frame (ORF) sequence of g1449 gene was predicted, and the length of the open reading frame was found to be 2670bp. Ustilago smut ( Ustilago esculenta ) haploid strain UET1 and Ustilago smut ( Ustilago esculenta ) The genomic DNA of the haploid strain UET2 was used as a template, and the primer Itd1-F / R was designed to amplify the Itd1 gene sequence, and the primer Itd1-UF / UR was amplified Itd1 The upstream promoter sequence of the gene, the primer Itd1-DF / DR amplifies the downstream promoter sequence of the Itd1 gene. The total RNA of the two stra...
Embodiment 2
[0029] Embodiment 2: wild rice smut ( Ustilago esculenta ) haploid strain UET1△Itd1 and Ustilago smut ( Ustilago esculenta ) Obtaining of haploid strain UET2△Itd1
[0030] 1. Cloning of upstream and downstream knockout vectors: Using the principle of homologous recombination, using the strain UET1 as a template, use primer Itd1-U2 / U3 to amplify the upstream knockout fragment, and use primer Itd1-D1 / D2 to amplify the downstream knockout fragment , using the plasmid pUM1507 as a template, use primer hyg-F / 3 to amplify the upstream segment of hygromycin, use primer hyg-4 / R to amplify the downstream segment of hygromycin, and fuse the upstream segment of the gene with the upstream segment of hygromycin PCR method, and use the primer Itd1-U2 / hyg3 to obtain the upstream knockout long fragment, connect it to the T vector, and transform Escherichia coli; use the method of fusion PCR for the downstream fragment of the gene and the downstream fragment of hygromycin, and use the prime...
Embodiment 3
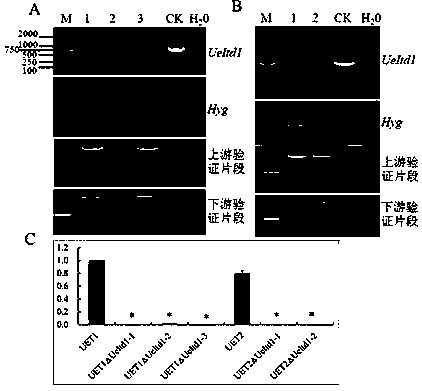
[0052] Embodiment 3: wild rice smut ( Ustilago esculenta ) haploid strain UET1△Itd1 and Ustilago smut ( Ustilago esculenta ) inoculation verification of haploid strain UET2△Itd1
[0053] 1) Shake the haploid strains of Ustilago smut UET1△Itd1 and UET2△Itd1 in YEPS liquid medium at 25-30℃ to OD 600nm 0.8-1.0, the cells were collected by centrifugation, and diluted with 0.5×YEPS liquid medium to a final concentration of OD 600nm 2.0-3.0 bacteria solution, mix the bacteria solution two by two for inoculation;
[0054] 2) Cultivate seedlings of the wild water pipe roots with more than 3 complete internodes, and cultivate them in the greenhouse for 15-20 days. Inoculate the tubular roots with more than 3 small seedling plants that have germinated, and pierce the base of the seedlings ;
[0055] 3) Inject the mixed bacterial liquid obtained in step 1) into the tubular root with a syringe until the bacterial liquid overflows;
[0056] 4) Trim the treated inoculated seedlings t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com