Method for detecting 3'-5' exo-activity of specific base by nuclease and kit thereof
A nuclease and kit technology, which is applied in the field of detecting the 3'-5' exocytosis activity of nucleases on specific bases, can solve the problems of limiting the scope of modified bases, complicated operation, long cycle, etc., to avoid radioactive contamination, Wide range of applications and flexible effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] Example 1. Establishment of a method for detecting the size of nuclease exonucleating activity to a specific base 3'-5'
[0049] 1. Detection principle
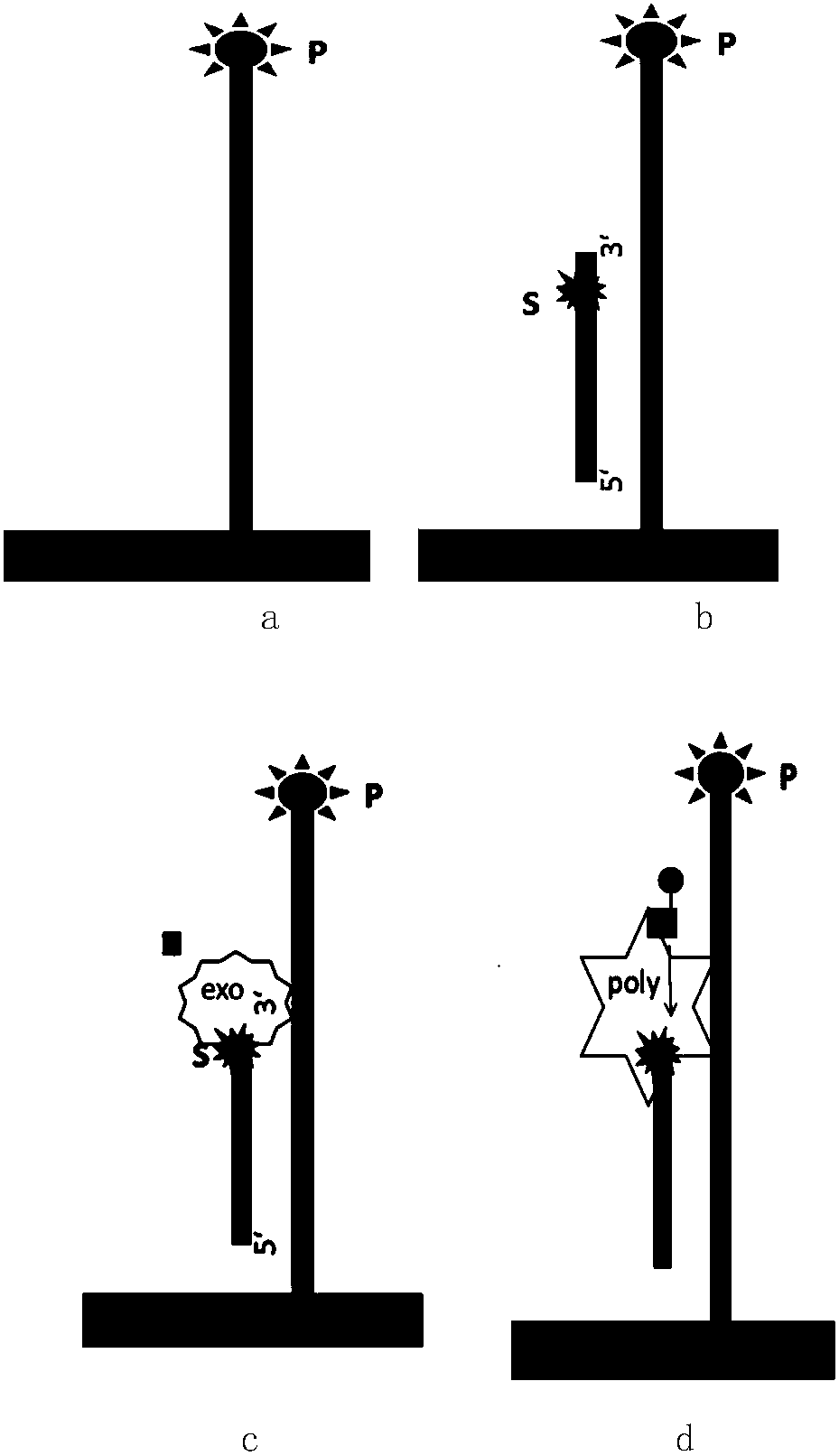
[0050] Such as figure 1 as shown,
[0051]1) Use a single-stranded DNA molecule immobilized on a solid phase with a known sequence as a template, such as oligonucleotides immobilized on magnetic beads with streptavidin and biotin or spotted on Oligonucleotides on the chip, etc., the exposed end of the single-stranded DNA molecule containing a known sequence needs to be modified to prevent exonuclease digestion, such as phosphorylation modification, sulfur modification, etc. (such as figure 1 a);
[0052] According to the experimental requirements, design a primer partially complementary to the known sequence, in which the phosphodiester bond between the penultimate base and the penultimate base in the order of 5'-3' is modified by sulfurylation , the 5' end is modified by phosphorylation, and the last base at the 3...
Embodiment 2
[0080] Example 2, detection of nuclease exonuclease activity on specific base 3'-5'
[0081] This embodiment uses exonuclease I as the internal reference nuclease of known activity (activity size is 10U / ul), detects the phi29DNA polymerase of Enzymatic and the phi29DNA polymerase of BGI according to the second method of embodiment 1 when there is no polymerization reaction In the case of the desired dNTP, the size of the 3'-5' exolytic activity of the unmodified C base under the condition of correct pairing. During the testing process, two control groups are set at the same time to prevent inaccurate results caused by the detected enzyme not cutting out the bases at all or cutting out all the bases on the chip.
[0082] Equipment used in the following method: BGISEQ-500 sequencer
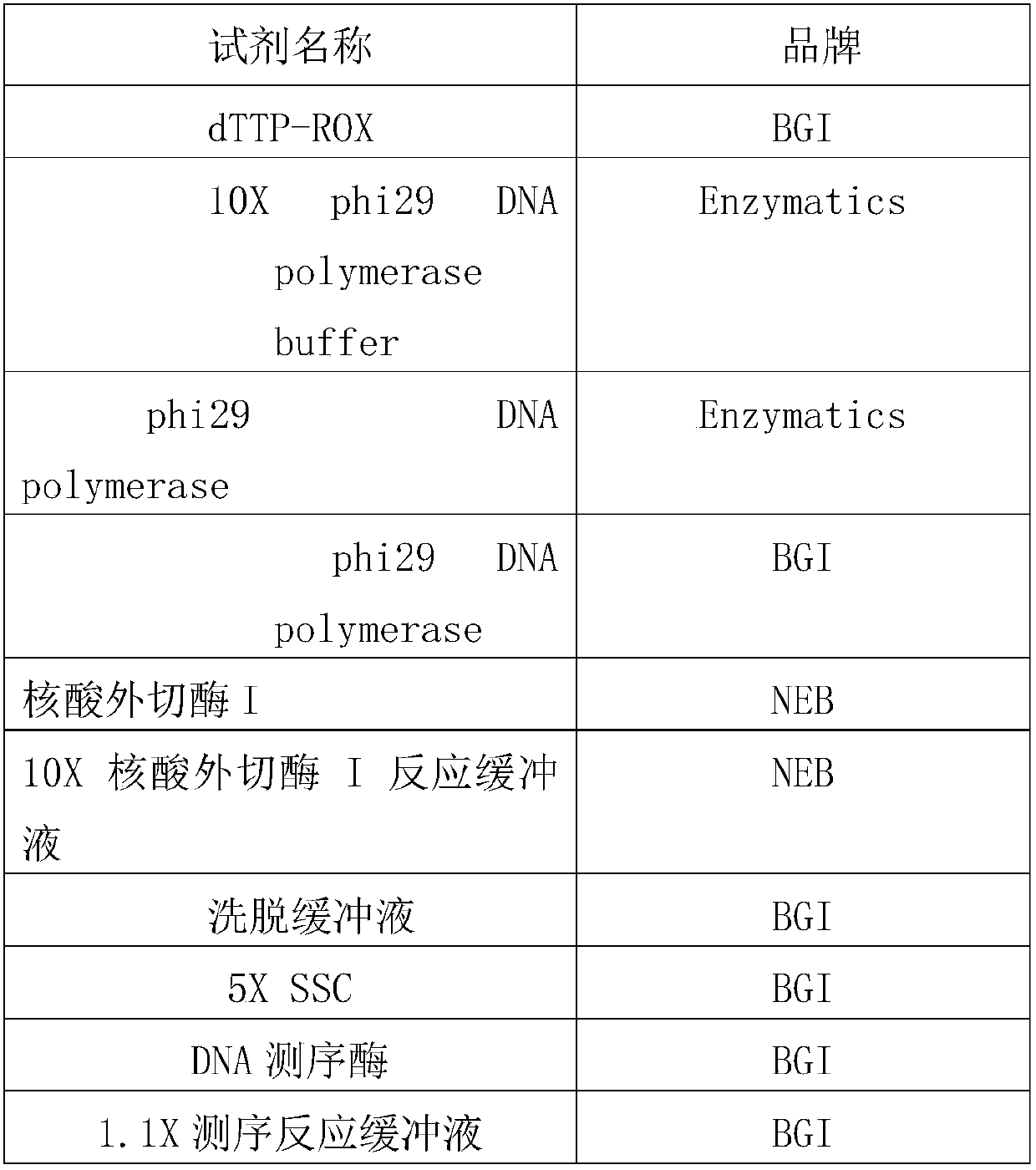
[0083] The reagents used in the following method are shown in Table 1 below:
[0084] Table 1
[0085]
[0086] Specific steps are as follows:
[0087] 1. Synthesize primers partially complem...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com