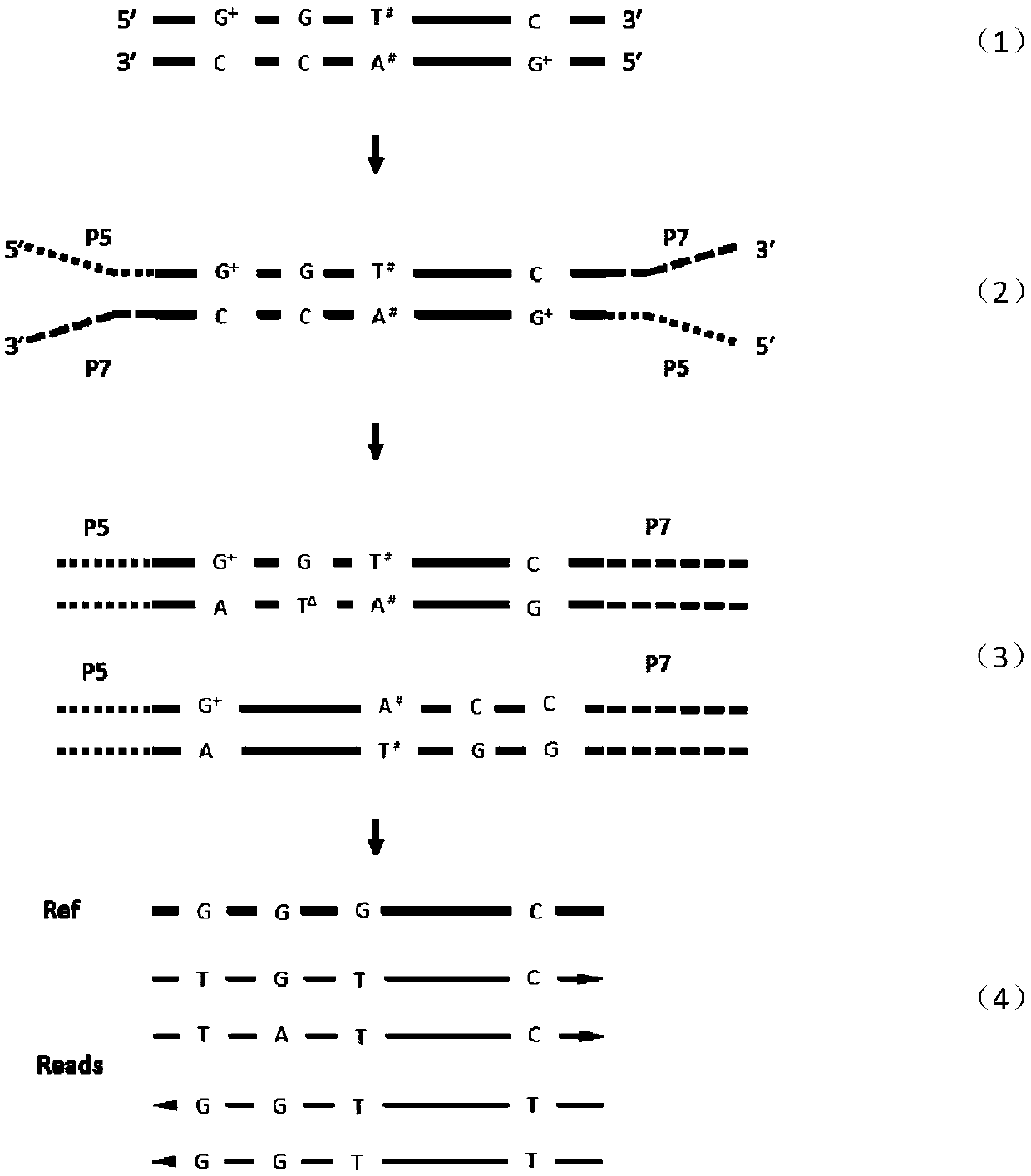
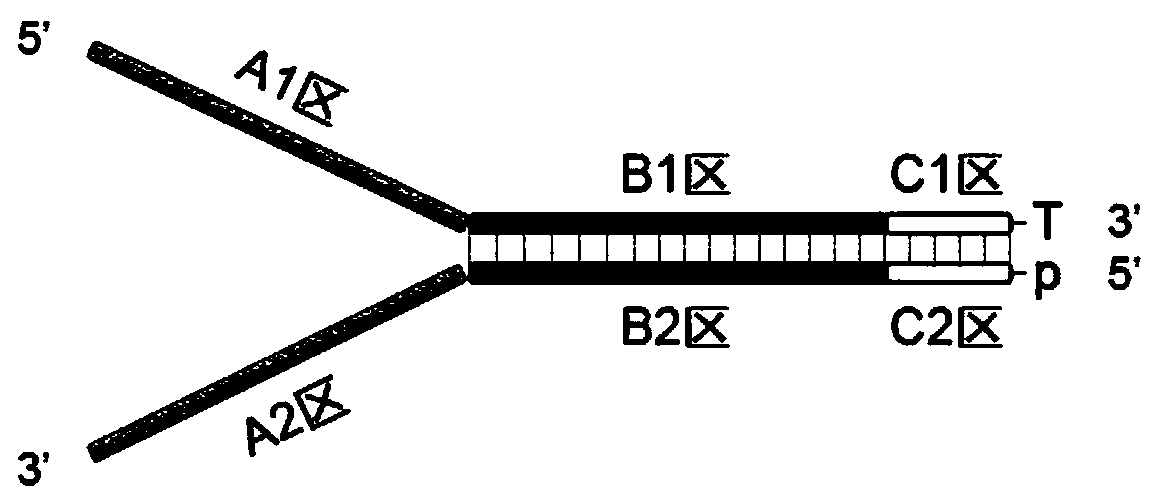
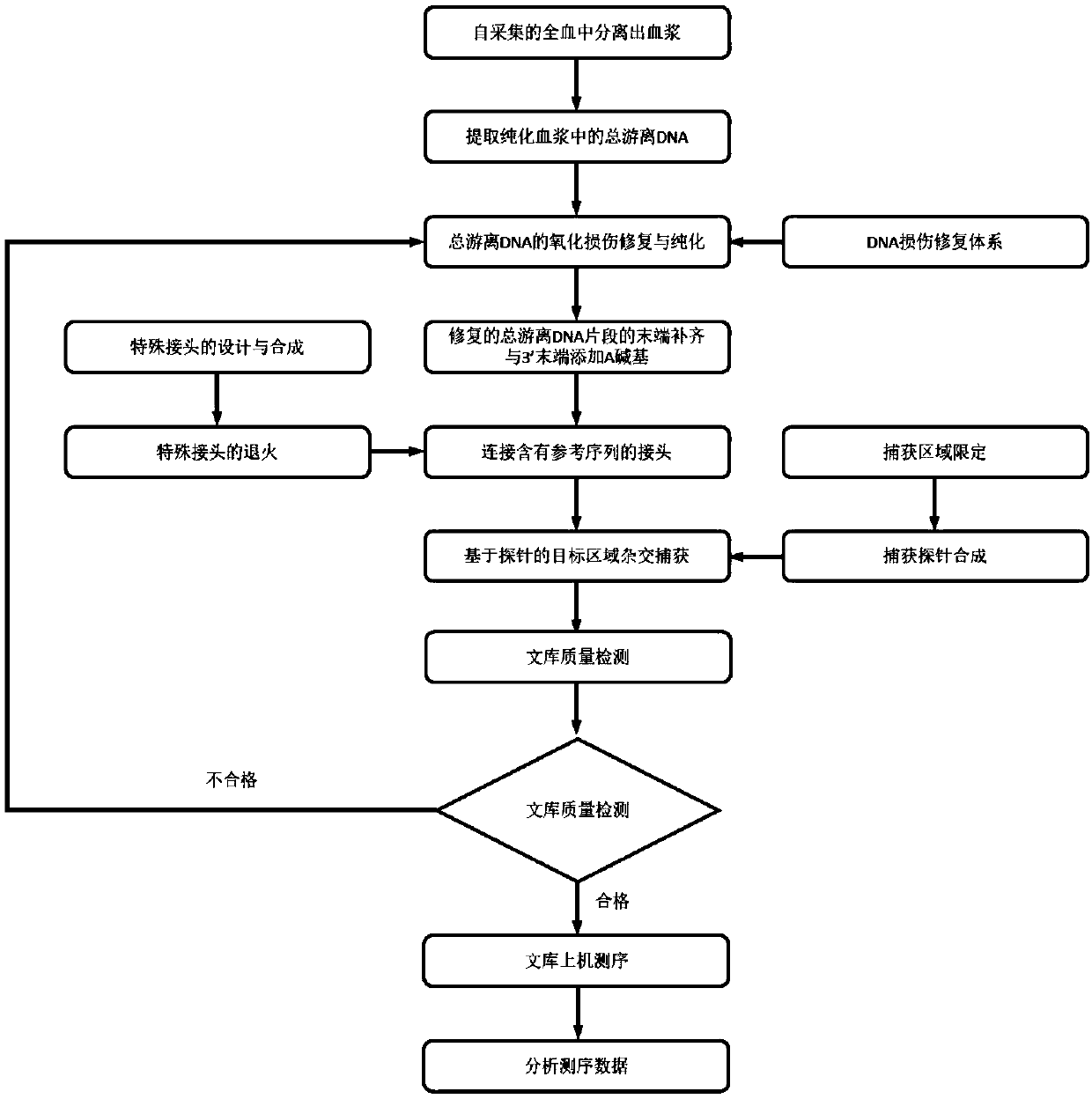
Detecting method of low-frequency DNA (Deoxyribonucleic Acid) mutation
A detection method and DNA library technology, applied in the field of detection of low-frequency DNA mutations, can solve problems such as detection data errors
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1D
[0057] Example 1DNA sample preparation
[0058] Take 30ml of whole blood from a patient with non-small cell lung cancer, and transport it at room temperature with 3 BCT tubes from Streck Company, and the transport time is about 46 hours.
[0059] Plasma was separated by a two-step centrifugation method, that is, the whole blood was centrifuged at 1600g for 10 minutes to obtain the supernatant; the supernatant of the previous step was centrifuged at 16000g for 10 minutes, and the obtained supernatant was plasma.
[0060] Two parts of the separated plasma were taken and immediately used the QIAamp Circulating Nucleic Acid Kit from Qiagen to extract the total free DNA respectively. The extraction amount is shown in Table 1.
[0061] Table 1
[0062] sample name
Embodiment 2
[0063] Example 2 DNA Oxidative Damage Repair
[0064] One of the DNA samples prepared in Example 1 was taken, and the DNA oxidative damage repair reaction mixture was prepared referring to the ratio in Table 2.
[0065] Table 2
[0066]
[0067] DNA repair buffer and enzyme mixture are FFPE DNA Repair Mix system of NEB Company.
[0068] The above system was transferred into a PCR machine, at 20° C. for 15 minutes, to repair the oxidative damage of DNA. Then, 186 μl of AMPure XP magnetic beads (purchased from Beckman) were added for purification, and 50 μl of nuclease-free water was used for elution.
Embodiment 3
[0069] Example 3 End completion of DNA fragments and addition of A bases at the 3' end
[0070] Take the total free DNA fragments repaired in Example 2, and prepare the reaction mixture according to the ratio in Table 3. In this example, KAPA Hyper Prep Kit was used for end repair and A buffer and end repair and A enzyme mixture.
[0071] table 3
[0072]
[0073] Move the above system into the PCR machine, and set up the program according to Table 4 to carry out the reaction.
[0074] Table 4
[0075]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com