Detection method of bacterial drug resistance gen and special kit thereof
A drug-resistant gene and kit technology, applied in the biological field, can solve problems such as difficult and accurate detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Embodiment 1, the acquisition of bacterial resistance gene and the design of detection primers
[0036] First, the acquisition of drug resistance genes
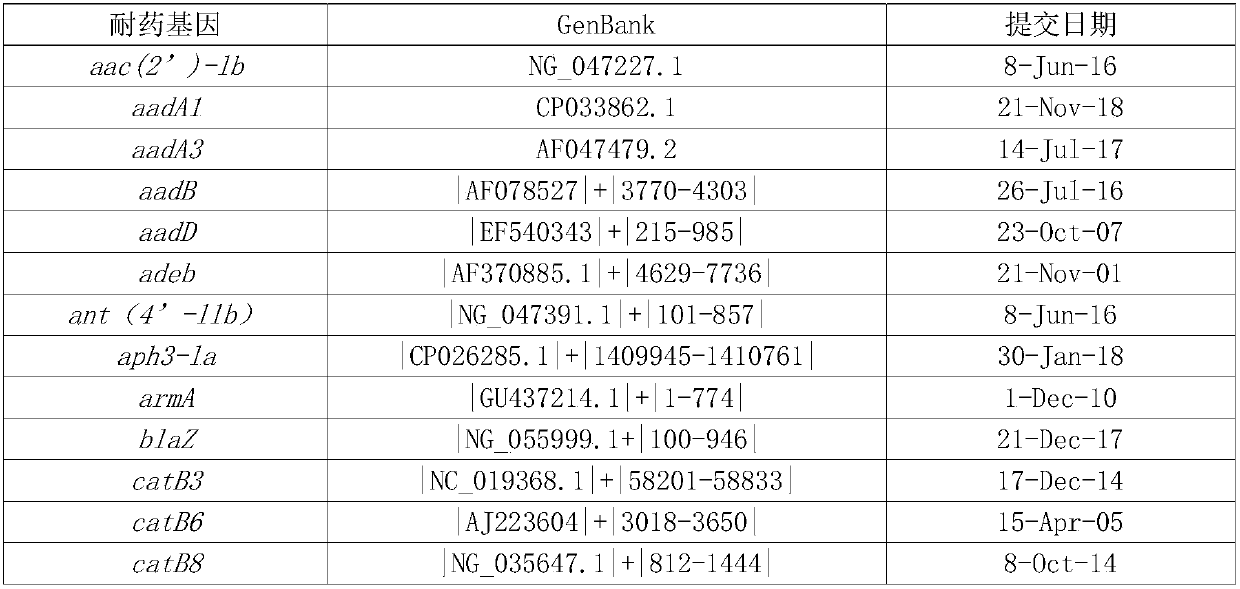
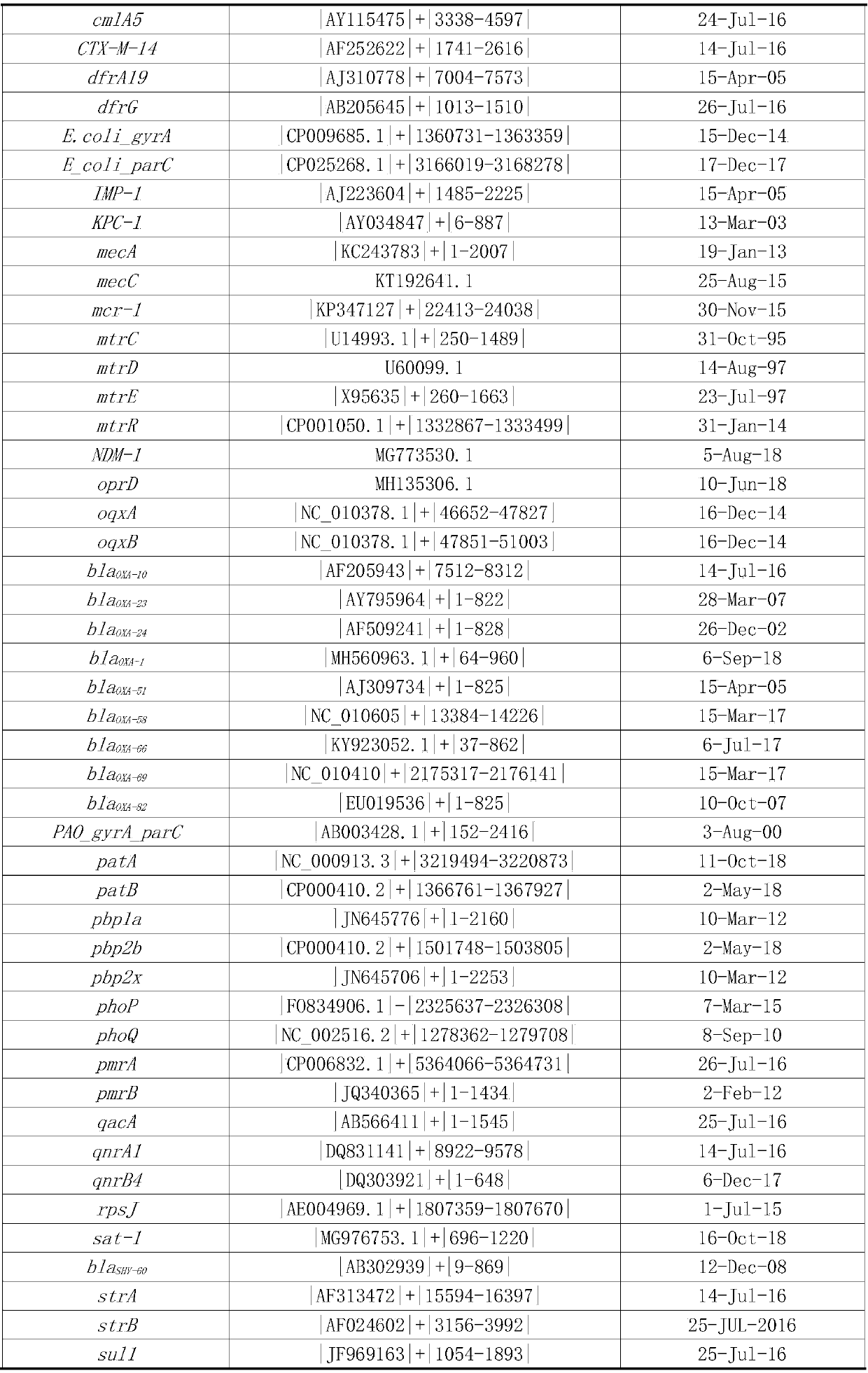
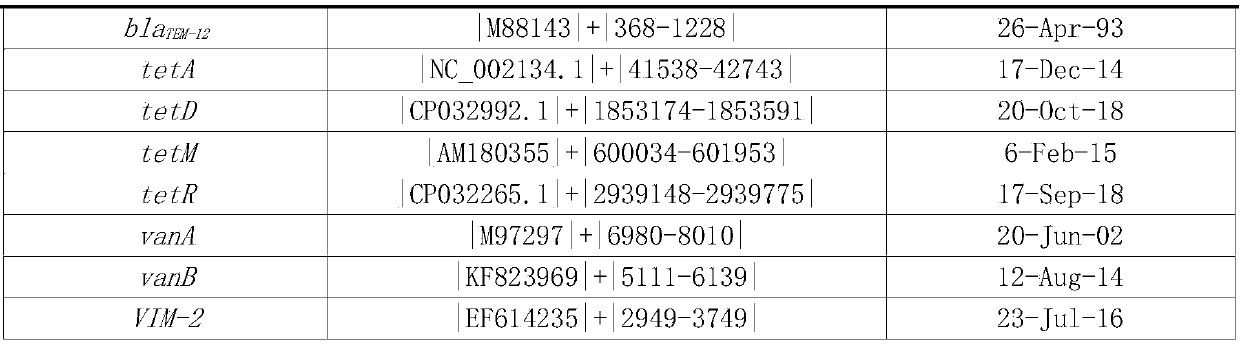
[0037] According to the types of commonly used clinical antibiotics and the types of common clinical pathogens, download from The Comprehensive Antibiotic Resistance Database (CARD; http: / / apcard.mcmaster.ca) and ARDB database (http: / / ardb.cbcb.umd.edu) The relevant fragments of drug resistance genes are sorted according to the types of pathogens, antibiotics, and drug resistance genes. The sorting method is as follows: The related fragments of drug resistance genes downloaded from ARDB database and CARD database are clustered by cd-hit, and 95% As the threshold, redundant sequences were removed; then BLAST was used to align the obtained fragments between species, and with 97% as the threshold, the specific fragments of drug resistance sites were screened to remove redundant sequences. Finally, a total of 68 drug resi...
Embodiment 2
[0054] Example 2. Application of primers for detecting bacterial resistance genes
[0055] (1) Detection of drug resistance genes of pathogens to be tested
[0056] 1. Extraction of pathogen nucleic acid
[0057] The nucleic acids of the pathogens to be detected in Table 4 were extracted by the magnetic bead method using Qiagen's MagAttract HMW DNA Kit. The specific extraction steps were carried out according to the instructions of the kit.
[0058] Table 4. Pathogens to be detected
[0059]
[0060]
[0061] 2. Construction of gene library
[0062] Library construction was performed using the Ion Ampliseq Library Kit 2.0 (Thermo Fisher Scientific, Cat. no. 4480441) based on the Ampliseq technology. Specific steps are as follows:
[0063] 1. Establish a DNA target amplification reaction with 2 primer pools
[0064] Ampliseq primers are divided into primer pool 1 (primer pool 1) and primer pool 2 (primer pool 2). The primers in primer pool 1 consist of the primers m...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com