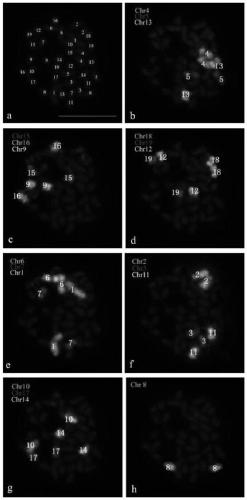
Method for identifying all chromosomes of poplar by using oligonucleotide probe
An oligonucleotide probe and oligonucleotide technology, which is applied in the field of identifying all the chromosomes of poplar by using oligonucleotide probes, can solve the problems such as the lag of cytogenetic research and the difficulty of accurately distinguishing chromosomes from chromosomes, and achieve the experimental result. Reproducible, high-resolution effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] A kind of method utilizing oligonucleotide probe to identify poplar whole chromosome, comprises the steps:
[0037] (1) Chromosome production
[0038] Collect young poplar root tips of 1 cm, immerse in 2mM 8-hydroxyquinoline solution for pretreatment at room temperature in the dark for 2.5 hours; rinse with water for 10 minutes, transfer to freshly prepared Carnot’s fixative, and store at -20°C until use ;
[0039] The root tip taken out of the fixative was rinsed in water for 10 minutes, the meristem part was cut with a double-sided blade, and the root tip meristem part was enzymatically hydrolyzed in an enzyme mixture of 4% cellulase and 2% pectinase at 37°C 1h; put the root tip meristem into deionized water to terminate the reaction;
[0040] Use a pipette to place the root tip on a clean glass slide, absorb excess water with a filter paper strip; mash the root tip tissue with tweezers and a dissecting needle; quickly add 20 μl of 60% glacial acetic acid dropwise, ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com