Mutant strains for realizing universal enzyme catalytic function diversity and construction method thereof
A construction method and mutant technology are applied in the field of mutant strains that realize the diversity of universal enzyme catalytic functions and the field of construction thereof, which can solve the problems of interspersed three-step dehydrogenation product streptavidin and the like, and achieve controllability and enrichment. The effect of functional information
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
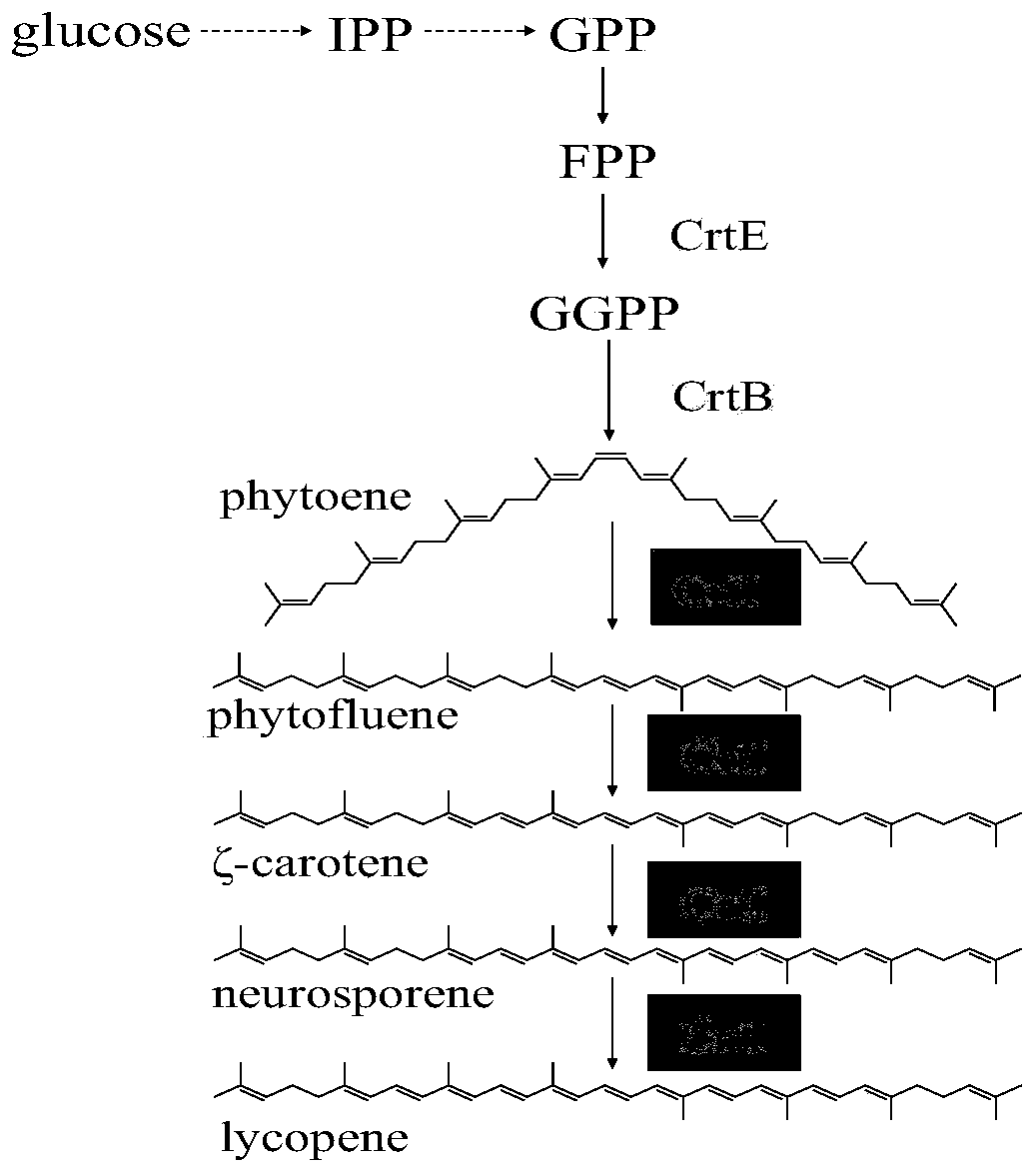
[0040] Example 1. Acquisition of Saccharomyces cerevisiae producing high phytoene
[0041] 1. Construction of modular integration plasmids
[0042] The Saccharomyces cerevisiae strain with high GGPP production, the strain number is SyBE_Sc14C10 (Chen Yan; Design, construction and fermentation process optimization of Saccharomyces cerevisiae with high lycopene production [D]; Tianjin University; 2017).
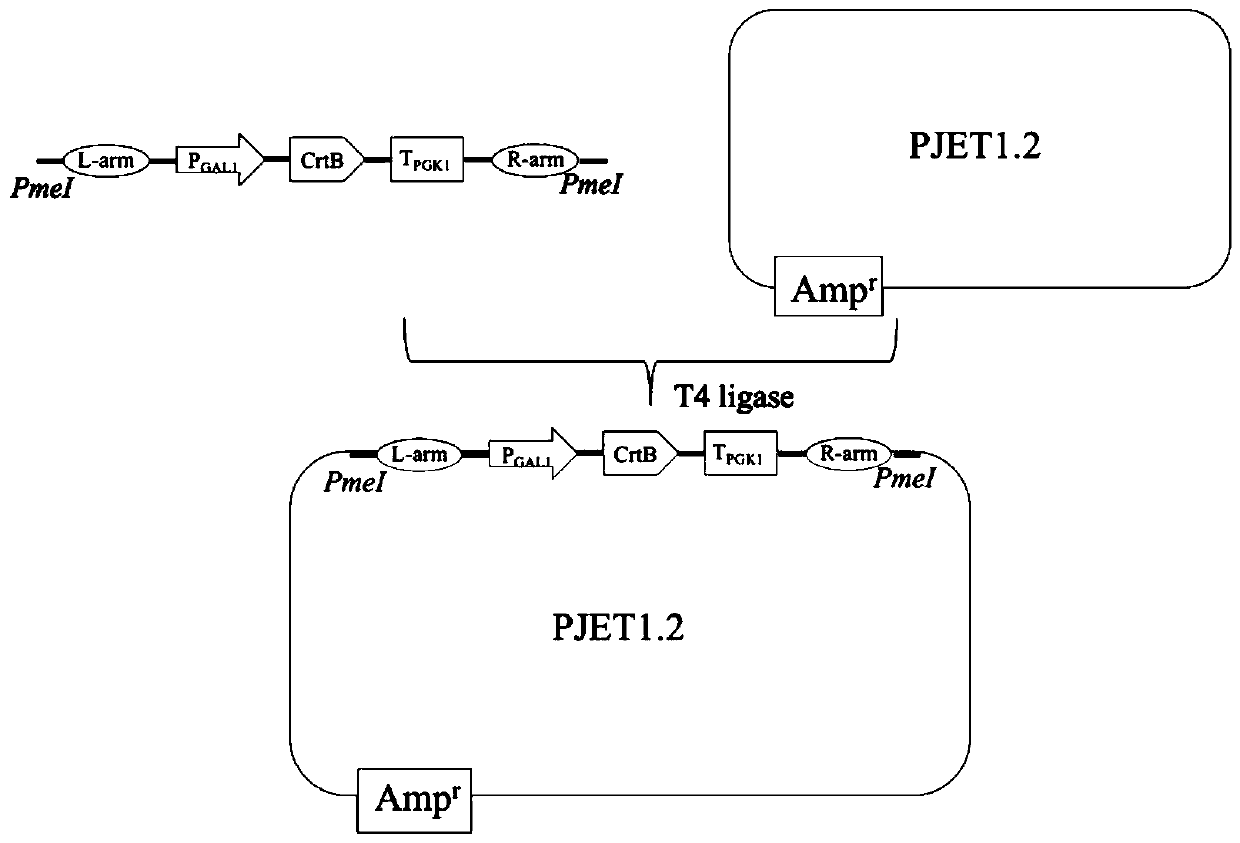
[0043] In order to realize the production of phytoene in Saccharomyces cerevisiae, elements such as yeast promoter Gal1-10, gene CrtB, terminator Pgk1t, upstream integrated homology arm, and downstream integrated homology arm were obtained by PCR amplification. CrtB was optimized according to the codons of Saccharomyces cerevisiae and was synthesized by Suzhou Jinweizhi Co., Ltd. after properly avoiding common restriction enzyme sites. The above elements were modularly assembled by overlapping extension PCR (OE-PCR) to obtain L-arm-P GAL1 -CrtB-Tpgk1-R-arm was named Module1 (S...
Embodiment 2
[0046] Embodiment 2, the construction of wild-type BtCrtI in Saccharomyces cerevisiae
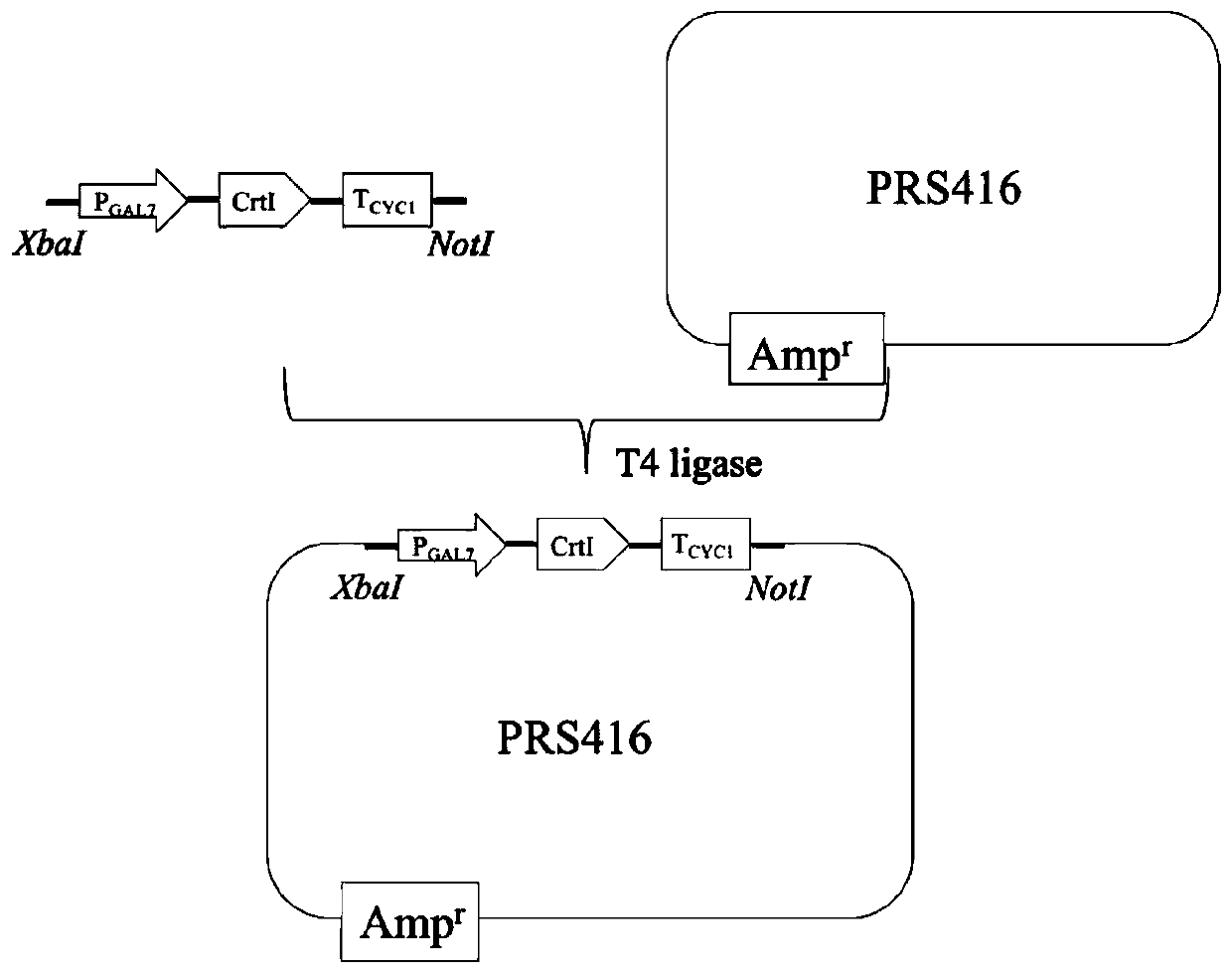
[0047] 1. Construction of a Saccharomyces cerevisiae strain that catalyzes phytoene to produce lycopene with CrtI ( image 3 )
[0048] The GAL7 promoter, the CYC1t terminator, and the gene BtCrtI synthesized by Suzhou Jinweizhi Company according to the codons of Saccharomyces cerevisiae, which were optimized according to the codon of S. The linear fragment of the NotI restriction site GAL7 promoter-BtCrtI-CYC1t terminator, that is, PGAL7-BtCrtI-TCYC1 (SEQ ID NO.2); Afterwards, the linear fragment and the PRS416 plasmid were treated with XbaI and NotI endonucleases , connected with T4 ligase, transformed into E. coli competent DH5α, screened by colony PCR, extracted plasmids for single and double enzyme digestion verification and sequencing verification to ensure that the target fragments are connected correctly and the base sequence is not mutated, so that the construction is correct rec...
Embodiment 3
[0057] Embodiment 3, the construction of wild-type BtCrtI in Saccharomyces cerevisiae
[0058] 1. Construction of BtCrtI mutation library by error-prone PCR
[0059] experiment material:
[0060] Strain SyBE_Sc04020001 with plasmid pRS416-BtCrtI
[0061] 10× error-prone PCR buffer: 70mm magnesium chloride hexahydrate, 500mm potassium chloride, 100mm tris, 0.1w / v gelatin
[0062] 10× error-prone PCR dNTPs: 10mm dCTPs and dTTPs, 2mm dGTPs and dATPs
[0063] 10mm manganese chloride tetrahydrate
[0064] The GAL7 promoter and CYC1t terminator were amplified by PCR, and the BtCrtI gene was amplified by random mutation of error-prone PCR using the plasmid containing BtCrtI as a template. The error-prone PCR system (100 μl) was configured as follows: add 10× PCR buffer 10 μl, 10× error-prone PCR dNTPs 10 μl, upper and lower primers 4 μl, template 2 μl, manganese chloride solution 5 μl, fasttaq enzyme 1 μl, and supplemented water 64 μl.
[0065] It should be noted that there is a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com