A nucleic acid aptamer of epithelial cell adhesion molecule epcam screened in human plasma and its preparation method and application
A technology for adhesion molecules and epithelial cells, applied in the fields of bioanalysis and clinical medicine, can solve problems such as poor affinity, and achieve low cost, improved capture efficiency, and high specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0074] Example 1 Screening of nucleic acid aptamers of epithelial cell adhesion molecule EpCAM in human plasma
[0075] 1) Chemically synthesize an initial DNA library of 80 bases, as shown in SEQ ID No.8, the two ends of the initial DNA library are fixed sequences, including a random sequence of 40 bases in the middle, which is 5'- AGC GTC GAA TAC CAC TACAG-40nt-CTA ATG GAG CTC GTG GTC AG-3' with a library capacity of 10 15above. Take 5nmol DNA initial library dissolved in binding buffer (12mmol / L PBS, pH 7.4, 150mmol / L NaCl, 5mmol / L KCl, 0.55mmol / L MgCl 2 ), perform denaturation treatment: 95°C for 10 minutes, place on ice for 5 minutes, and then place at room temperature for 10 minutes;
[0076] 2) In the environment of human plasma, incubate the processed DNA initial library with Ni agarose microspheres at 37°C, carry out reverse screening, and collect the liquid that is not combined with Ni agarose microspheres; among them, the human plasma The preparation method is as...
Embodiment 2
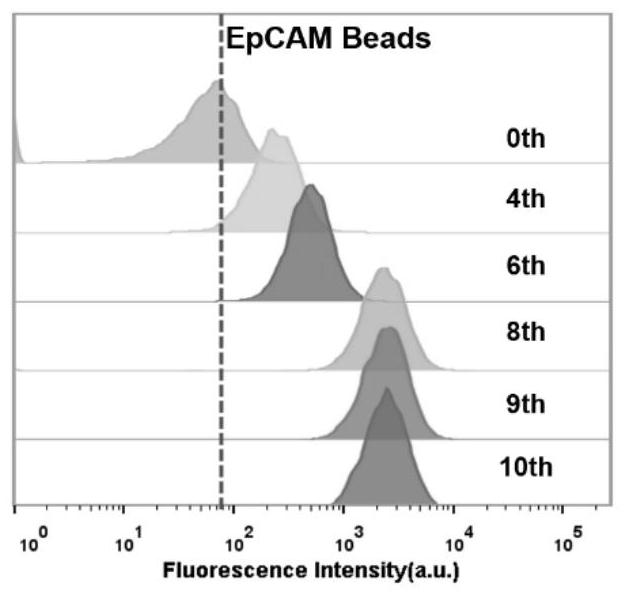
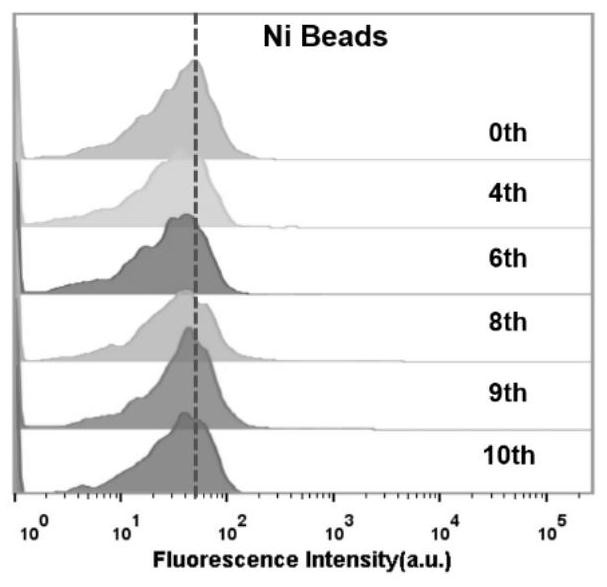
[0081] Example 2 Investigating the Binding Ability of Single-Stranded DNA Enrichment Library and EpCAM Protein by Flow Cytometry
[0082] First, PCR amplify the 0, 4, 6, 8, 9, and 10 rounds of DNA enrichment libraries with fluorescent labels, using the forward primer (SEQ ID No.9): 5'-FAM-AGC GTC GAA TAC CAC TAC AG-3' and reverse primer (SEQ ID No.10): 5'-Biotin-CTG ACC ACG AGC TCC ATT AG-3', pre-denaturation at 94°C for 3 minutes, 94°C for 30s, 53°C for 30s, 72°C for 30s, Amplify for 10 cycles, and finally extend at 72°C for 5 min; the PCR product is double-stranded DNA with FAM at the 5' end and biotin at the 3' end, add streptavidin agarose microspheres, react at room temperature for 30 min, and then Use 0.1mol / L NaOH to carry out alkali denaturation and single-strandization, desalt and filter through NAP-5 column, and purify to obtain single-stranded DNA enrichment libraries for the 0th, 4th, 6th, 8th, 9th, and 10th rounds;
[0083] Use 100 μL of human plasma to prepare t...
Embodiment 3
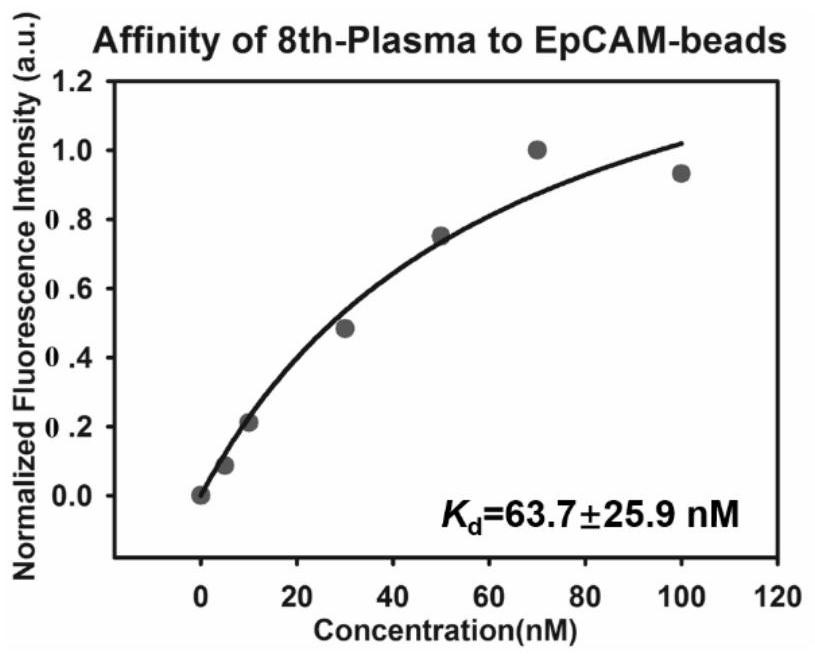
[0085] Example 3 Determination of the dissociation constant between the eighth round of single-stranded DNA enrichment library and EpCAM protein obtained by flow cytometry
[0086] First, PCR amplifies the 8th round DNA enrichment library with fluorescent labeling, using the forward primer 5'-FAM-AGC GTCGAA TAC CAC TAC AG-3' (SEQ ID No.9): with the reverse primer (SEQ ID No. .10): 5'-Biotin-CTG ACCACG AGC TCC ATT AG-3', the PCR product is double-stranded DNA with FAM at the 5' end and biotin at the 3' end, add streptavidin agarose micro spheres, reacted at room temperature for 30 minutes, and then used 0.1mol / L NaOH to carry out alkali denaturation and single-strandization, desalted and filtered through NAP-5column, and purified to obtain the eighth round of single-stranded DNA enrichment library;
[0087] Use 0,5,10,50,75,100nmol / L concentration gradient of the eighth round of single-stranded DNA enrichment library and EpCAM protein agarose microspheres to determine the disso...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com