Nitrilase xinit1 and its coding gene and application
A technology of nitrilase and gene, applied in nitrilase XiNit1 and its coding gene and application fields, to achieve the effects of high tolerance, wide reaction pH and high enzyme activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
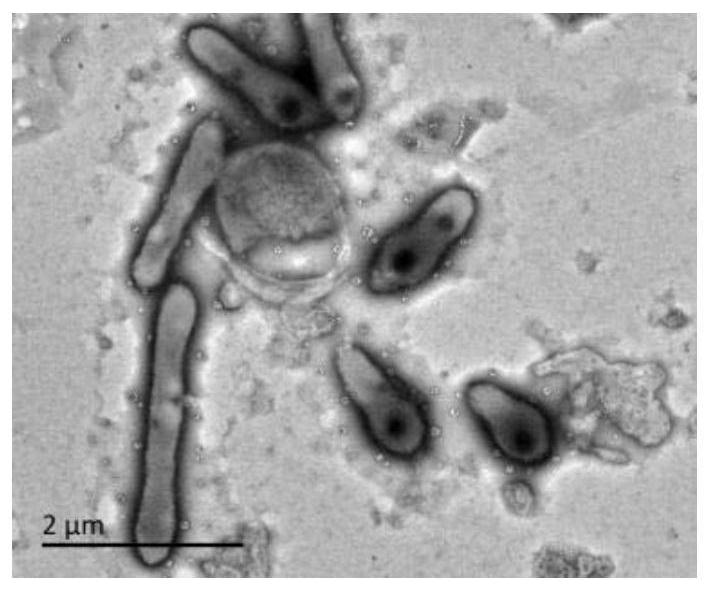
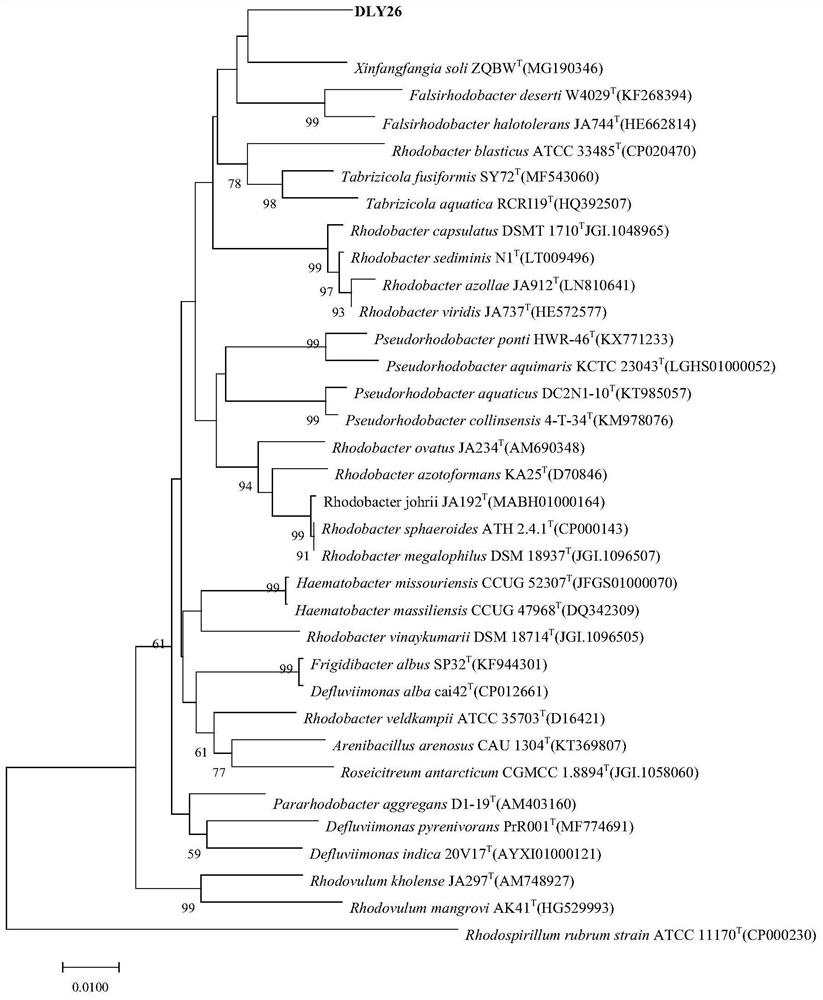
[0045] Example 1, Isolation, Identification and Preservation of Xingfangfangbacterium DLY26
[0046] 1. Separation
[0047] Take about 1ml of activated sludge sample (taken from the petrochemical refinery sewage treatment system), and dilute to 10 times volume, 20 times volume, 50 times volume, 100 times volume and 1000 times volume in turn with sterile distilled water. Spread 100 μl of the diluted sample on the surface of the solid medium by the coating plate method, incubate at 30°C, and observe the growth of the colonies on the surface every day. Pick colonies with different colors and shapes, and purify and culture them by the three-section line method until a single colony with the same shape and size can be observed on the surface of the culture medium.
[0048] 2. Identification
[0049] The purified bacterial strains were inoculated on TSA plates, cultivated at 30°C for 24h, and then observed the morphology, size and other characteristics of the cells using a transmi...
Embodiment 2
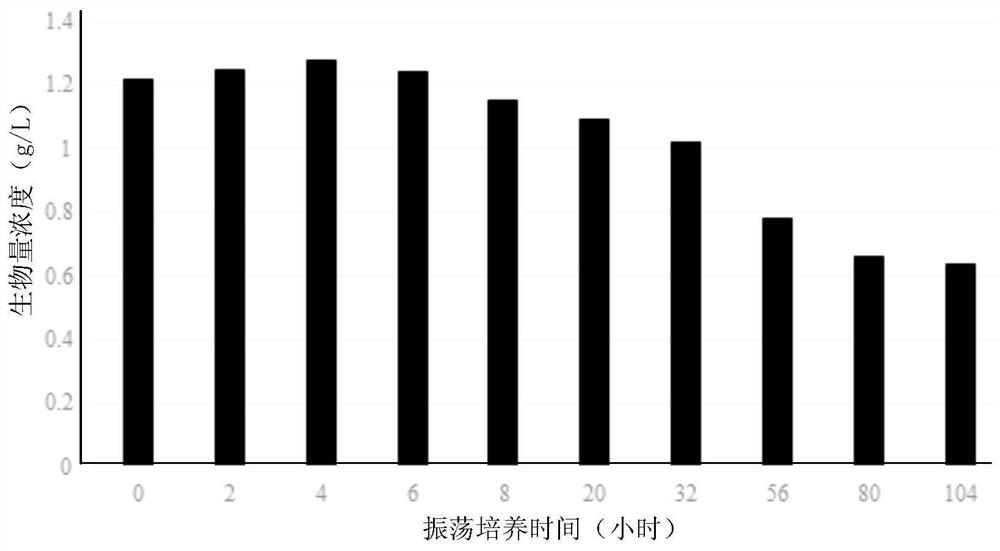
[0066] Embodiment 2, the degradation action of Xinfangfang bacteria DLY26 to acrylonitrile
[0067] Na 2 HPO 4 / NaH 2 PO 4 The formula of the buffer solution: disodium hydrogen phosphate 14.2g, sodium dihydrogen phosphate 12.2g, dissolved in 800mLddH 2 O, with ddH 2 O was adjusted to 1L, and the pH was adjusted to 7.0.
[0068] The formula of the activation medium: tryptone soybean broth medium (TSB) 30g, with ddH 2 O was dissolved and the volume was adjusted to 1L, and the pH was adjusted to 7.0.
[0069] Basal medium: 2.0g disodium hydrogen phosphate, 1.0g potassium dihydrogen phosphate, 0.5g yeast powder, 0.2g magnesium sulfate, 0.03g ferrous sulfate, dissolved in 800mL Na 2 HPO 4 / NaH 2 PO 4 buffer, then add 3.25mL of glycerol and mix well, then use Na 2 HPO 4 / NaH 2 PO 4 Make up to 1L of buffer.
[0070] 1. Preparation of seed solution
[0071] Inoculate Xingfangfang bacteria DLY26 into the activation medium, shake culture at 30°C and 150rpm, and obtain se...
Embodiment 3
[0091] Embodiment 3, the preparation of nitrilase (XiNit1 protein)
[0092]After a large number of sequence analysis, alignment and functional verification, a new protein was found from DLY26 of Xingfangfang bacteria, which was named as XiNit1 protein, as shown in sequence 1 of the sequence table. The gene encoding XiNit1 protein in Xingfangxiang bacteria DLY26 is named as xinit1 gene, and its coding frame is shown in sequence 2 of the sequence list.
[0093] 1. Construction of recombinant plasmids
[0094] 1. Using the genomic DNA of Xingfangfang bacteria DLY26 as a template, PCR amplification is carried out with primer pairs composed of DN1-F and DN1-R, and the PCR amplification products are recovered.
[0095] DN1-F: 5'- CACC ATGCTGGCGGAAGG-3';
[0096] DN1-R: 5'-CGGCCAGCTCGTCATGAC-3'.
[0097] 2. Take the PCR amplification product obtained in step 1 and connect it to the pDE1 vector to obtain the recombinant plasmid pDE1-xinit1.
[0098] The pDE1 vector (pDE1Vector) ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com