Method for preparing DNA self-assembly electrochemical biosensor through utilizing DSN and DNAzyme
A biosensor, electrochemical technology, applied in the field of electrochemical detection, can solve the problems of long time consumption, low detection sensitivity, limited qRT-PCR application, etc., and achieve the effect of high selectivity, broad application prospects and high specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Example 1. Detect whether the sample to be tested contains microRNA-141
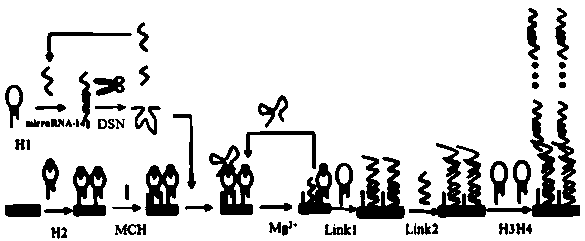
[0035] figure 1 Schematic diagram of the construction process of the present invention
[0036] The following example uses microRNA-141 as the microRNA to be tested, and the nucleotide sequence of microRNA-141 is UAACACUGUCUGGUAAAGAUGG.
[0037] 1. Samples to be tested
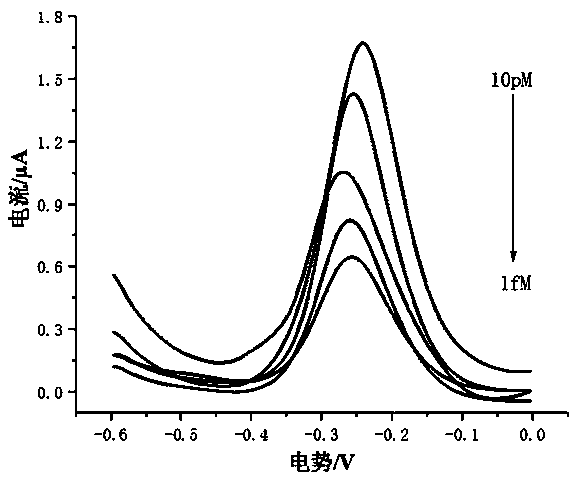
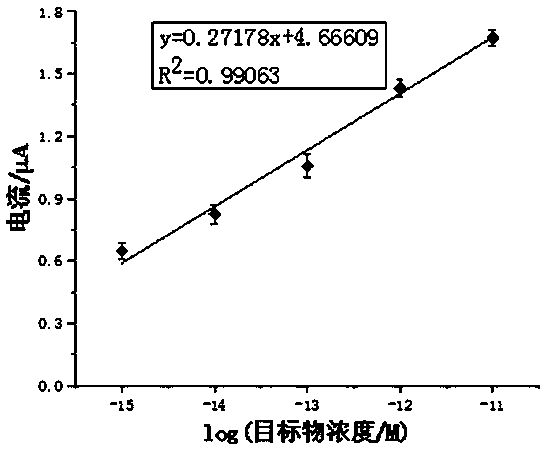
[0038] The embodiment of the present invention uses a series of solutions with a concentration of 1 fM to 10 pM as the sample to be tested. The sample to be tested in the present invention can also be derived from plasma or serum. The specific method is as follows:
[0039] All electrochemical tests are carried out on the CHI660C electrochemical workstation. The three-electrode system includes: gold electrode (working electrode) that completes DNA self-assembly, platinum wire electrode (counter electrode), and silver-silver chloride (Ag / AgCl) reference electrode. The electrochemical detection was carried out in RuHex solution. First, let...
Embodiment 2
[0067] In order to evaluate the specificity of the method of the present invention, several microRNAs with different sequences (including (a) blank (b) microRNA-200a (sequence UAACACUGUCUGGUAACGAUGU), (c) microRNA-429 (sequence UAAUACUGUCUGGUAAAACCGU) were analyzed with the same experimental procedure in Example 1. ), (d) single-base mismatch microRNA-141 (sequence UAACACUGUCUCGUAAAGAUGG), (e) microRNA-141. The concentration of microRNA-141 is 10pM for detection.
[0068] Test results such as Figure 4 As shown, the electrochemical signal generated by 10pM microRNA-141(e) is significantly higher than other samples. The results show that this method has high sequence specificity and is expected to be used to identify different microRNA sequences.
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com