Mutant heparinase I, encoding nucleotide sequence thereof, recombinant carrier and host cell with nucleotide sequence and application
A technology of nucleotide sequence and recombinant vector, applied in the field of molecular biology, can solve the problems of poor stability of heparinase I, decrease, and the yield is difficult to exceed 10%, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0050] According to one aspect of the present invention, a method for preparing mutant heparanase I, the preparation method comprises the following steps:
[0051] (a), first synthesizing the nucleotide sequence encoding the above-mentioned mutant heparanase I, and then combining the nucleotide sequence with the eukaryotic recombinant expression vector to obtain the recombinant vector;
[0052] (b) Transforming the recombinant vector into host cells, followed by inducing expression to obtain mutant heparanase I.
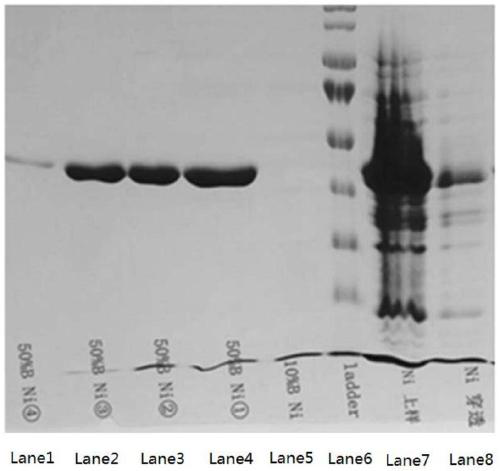
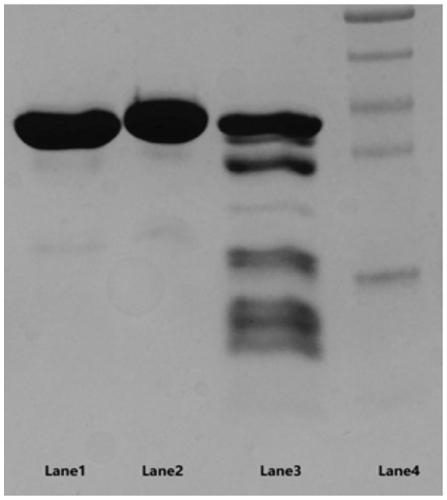
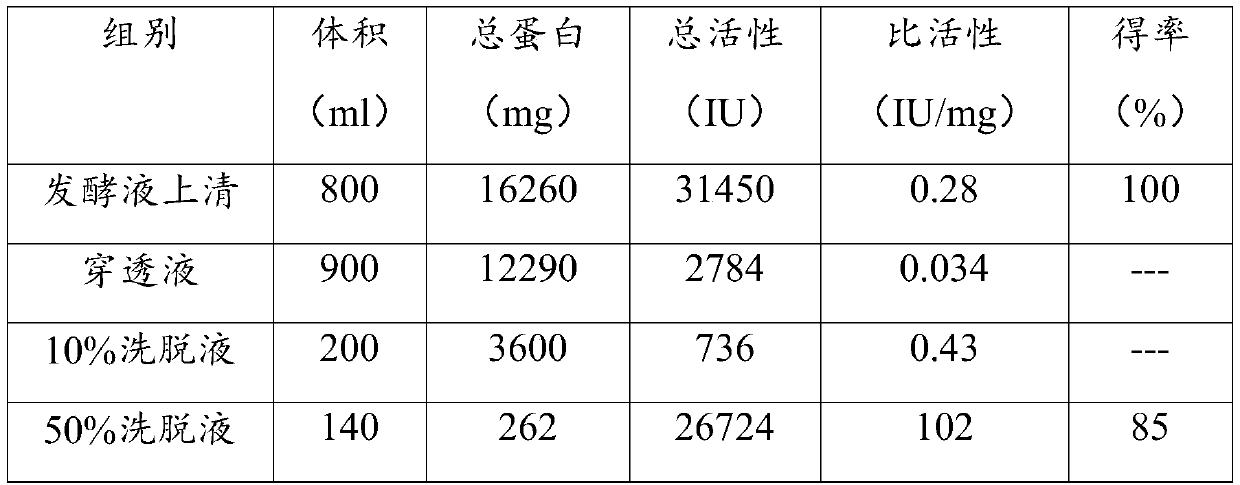
[0053] The preparation method of the mutant heparanase I provided by the present invention, the preparation method combines the synthesized nucleotide sequence encoding the above mutant heparanase I with the eukaryotic recombinant expression vector to obtain a recombinant vector by means of genetic engineering , and transfer the recombinant vector into host cells to induce expression, and obtain mutant heparanase Ⅰ. The preparation method can produce mutant heparana...
Embodiment 1
[0057] Example 1 Synthesis of Mutant Heparanase I Nucleotide Sequence and Construction of Expression Vector
[0058] (a), search the UniProt library to obtain the protein sequence of Flavobacterium heparinus heparanase I (AccessionNo.Q05819), and enter the sequence into the codon bias analysis tool Gene designer software;
[0059] (b) According to the codon usage preference of Pichia pastoris in the Pichia pastoris codon preference data table, the protein sequence is reverse-translated into a DNA sequence, so that the codons of the DNA sequence are all preferred by Pichia pastoris (using online codon optimization tool for verification). At the same time, we input the protein sequence from amino acid 22 to amino acid 384 of Q05819 into the online tool "PeptideCutter" to predict potential protease cleavage sites;
[0060] (c) Perform mutation transformation on potential protease cleavage sites, specifically, site-directed mutation of Gln at position 2 of the existing heparanase...
Embodiment 2
[0063] The electrotransformation of embodiment 2 expression vector
[0064] 1. First prepare GS115 competent cells:
[0065] (1) Pick a single colony of yeast GS115 from the YPD plate, inoculate it in 5ml of YPD liquid medium, and culture overnight at 29°C with shaking at 180rpm; inoculate 1mL of the overnight cultured GS115 bacterial liquid into 100mL of YPD medium at 29°C , Shake culture at 180rpm until the OD600 value is about 1.3-1.5;
[0066] (2) Take 50mL of GS115 bacteria solution, centrifuge at 4°C, 4000rpm for 5min to collect the bacteria, discard the supernatant, and absorb the residual liquid; add 40mL of ice-cold sterile water to wash the bacteria, and centrifuge at 4°C, 4000rpm for 5min , discard the supernatant; and repeat this step once;
[0067] (3) Add 10 mL of ice-cold sterile water to wash the cells, centrifuge at 4° C. 4000 rpm for 5 min, discard the supernatant; resuspend the cells with 2 mL of ice-pre-cooled 1 M sorbitol, and centrifuge at 4° C. 4000 rp...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com