A single quantum dot fluorescent nanosensor based on enzyme-free catalyzed self-assembly and its preparation method and application
A fluorescent nanometer, non-enzyme-catalyzed technology, applied in the field of biological analysis, can solve the problems of reducing detection accuracy, complicating the design of the scheme, increasing the detection cost, etc., and achieves the advantages of less sample consumption, improved sensitivity, and reduced false positive signals Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
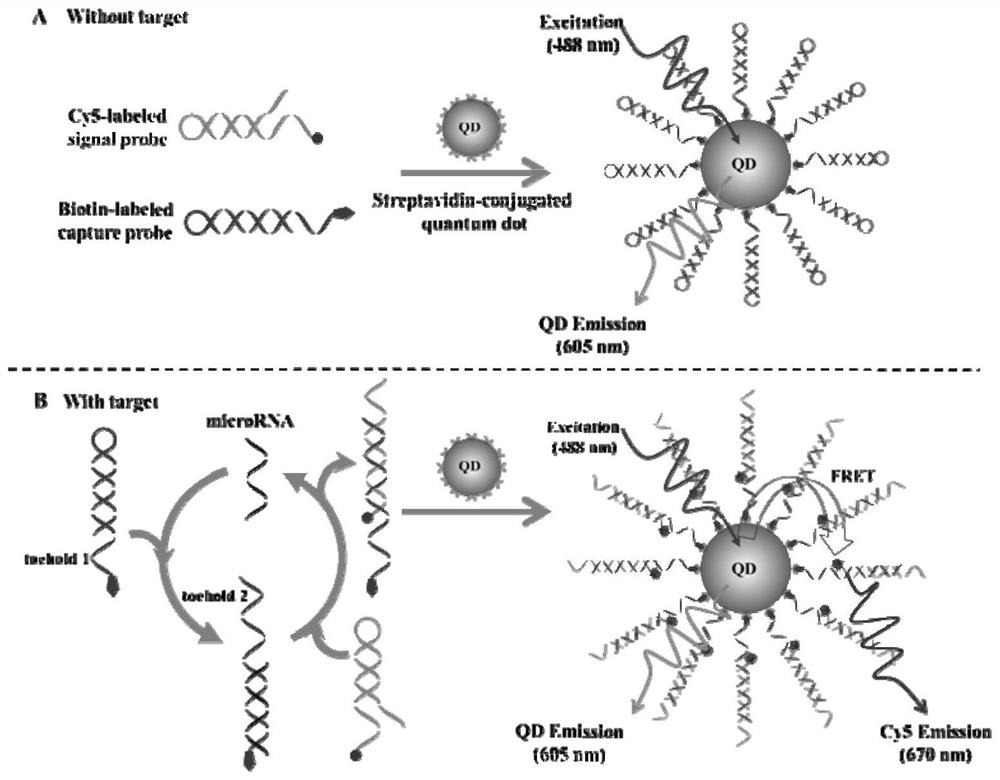
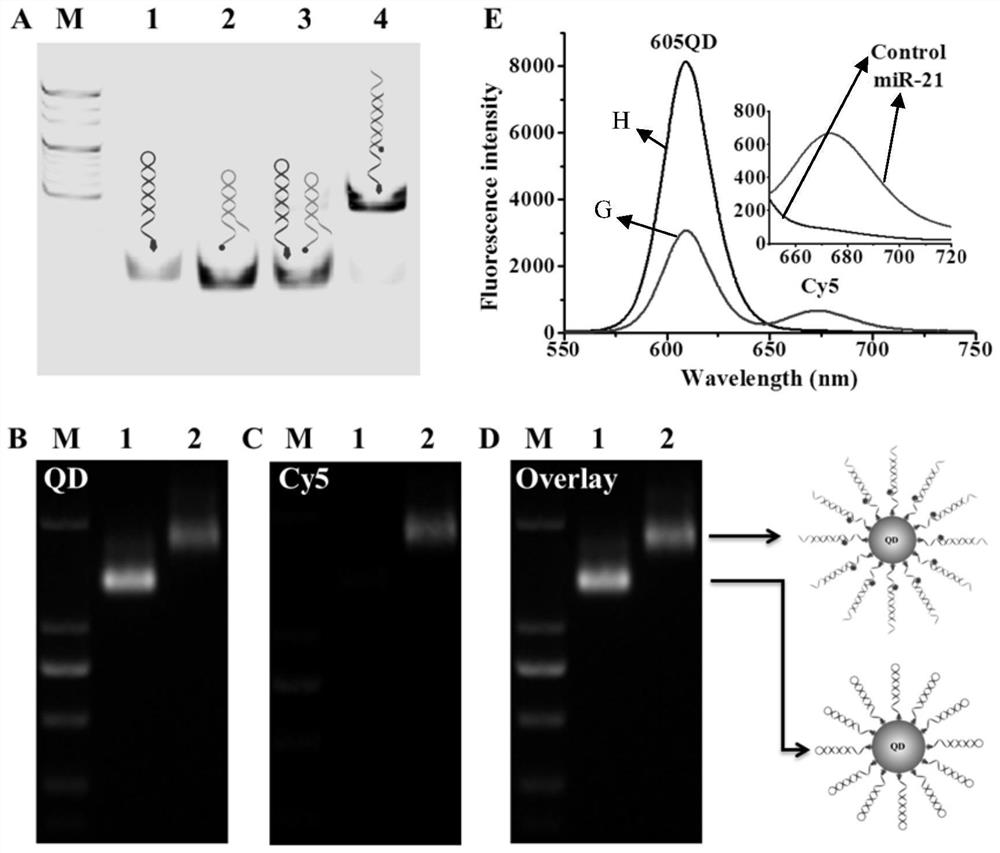
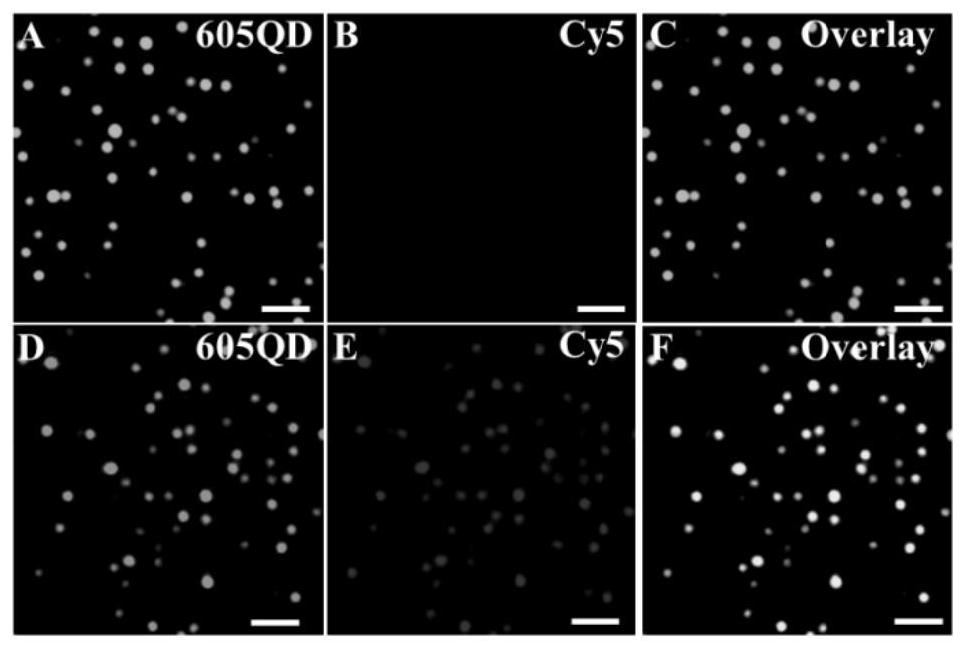
[0045] Detection of circulating microRNAs and formation of QD / DNA / Cy5 nanocomplexes: The detection of circulating microRNAs consists of two sequential steps: first, signal probes and capture probes were mixed in 1× reaction buffer (20 mM per liter of triplicates). hydroxymethylaminomethane-hydrochloric acid, 140 mM NaCl, 5 mM KCl, pH 7.5) diluted to 2 μM per liter, then incubated at 95°C for 5 min, and Slowly cool to room temperature to form hairpin structure. The prepared hairpin probes were used immediately or kept at -20°C for later use. Second, the 60 μl reaction system includes the indicated concentration of miR-21, 5.4 μl capture probe (2 μmol per liter), 5.4 μl signal probe (2 μmol per liter), 6 μl 10× reaction Buffer (200 mmol / L Tris-HCl, 1.4 mmol / L NaCl, 50 mmol / L KCl, pH 7.5, react at 37°C for 3 hours, then add 6 Microliters of 605QDs (50 nanomoles per liter) were incubated at 37°C for 10 minutes to form 605QD / DNA / Cy5 nanostructures.
[0046] Polyacrylamide gel el...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com