Amino acid dehydrogenase and application thereof
An amino acid dehydrogenase, dehydrogenase technology, applied in the directions of enzymes, oxidoreductases, enzymes, etc., can solve the problems of serious environmental pollution, complex reaction process, harsh reaction conditions, etc., and achieve high optical purity, simple equipment, and operation. handy effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] To construct a single-point mutation (G60S) amino acid dehydrogenase mutant, to mutate the 60 amino acid, the specific steps are as follows:
[0029] (1) Introducing mutations: Design primers based on the nucleotides shown in SEQ ID NO. 02, and design forward and reverse primers containing different positions. The forward and reverse primers are as follows:
[0030] Upstream primer (SEQ ID NO.05): G60S-F GTGCTGAGATTGTCAAAATCAATG
[0031] Downstream primer (SEQ ID NO.06): G60S-R TGATTTTGACAATCTCAGCACATC
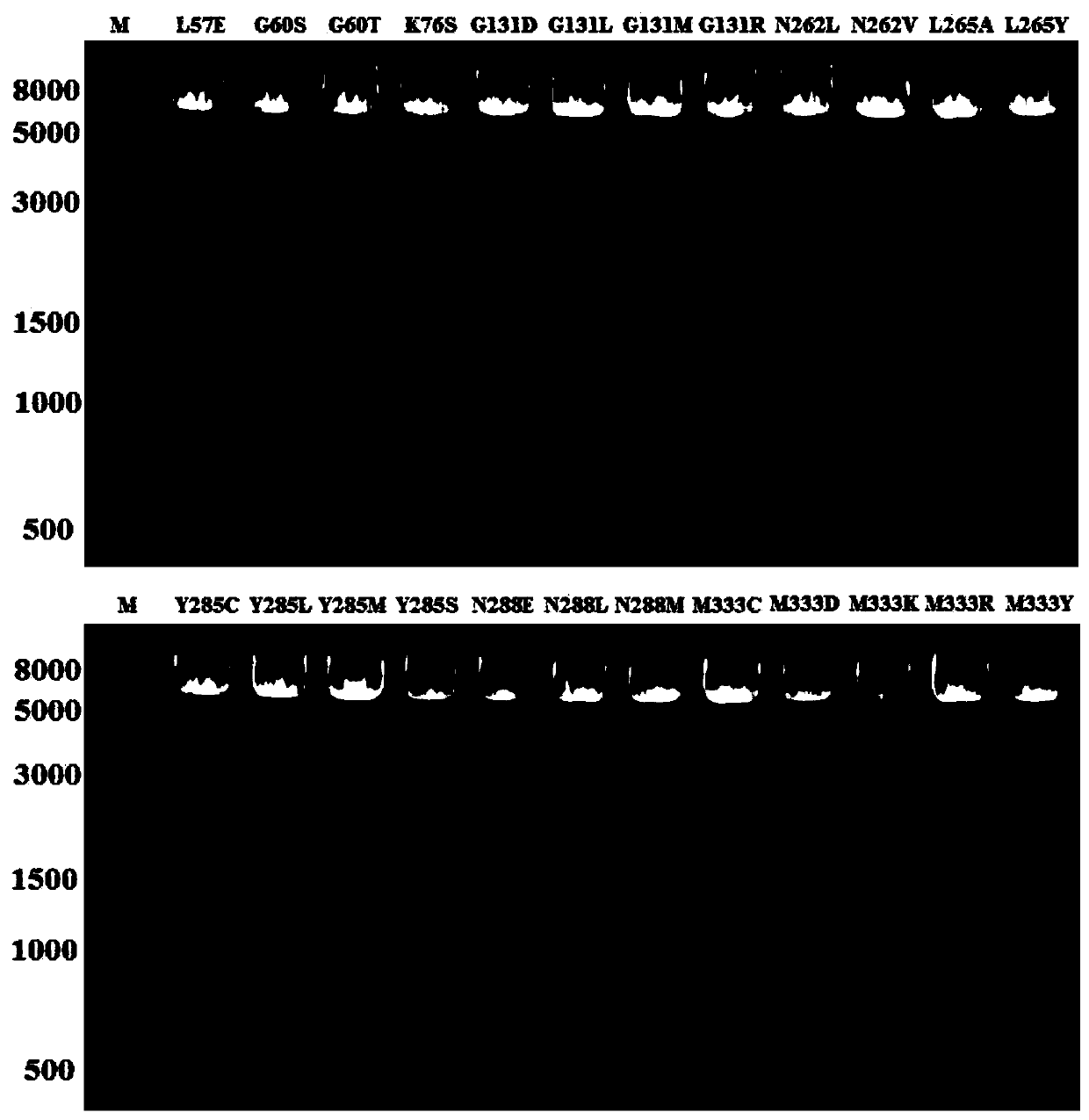
[0032] After mixing the primers and the template plasmid, add high-fidelity Taq polymerase KOD-Plus to perform PCR amplification of the whole plasmid. After PCR, the PCR products are detected by electrophoresis, such as figure 1 Shown.
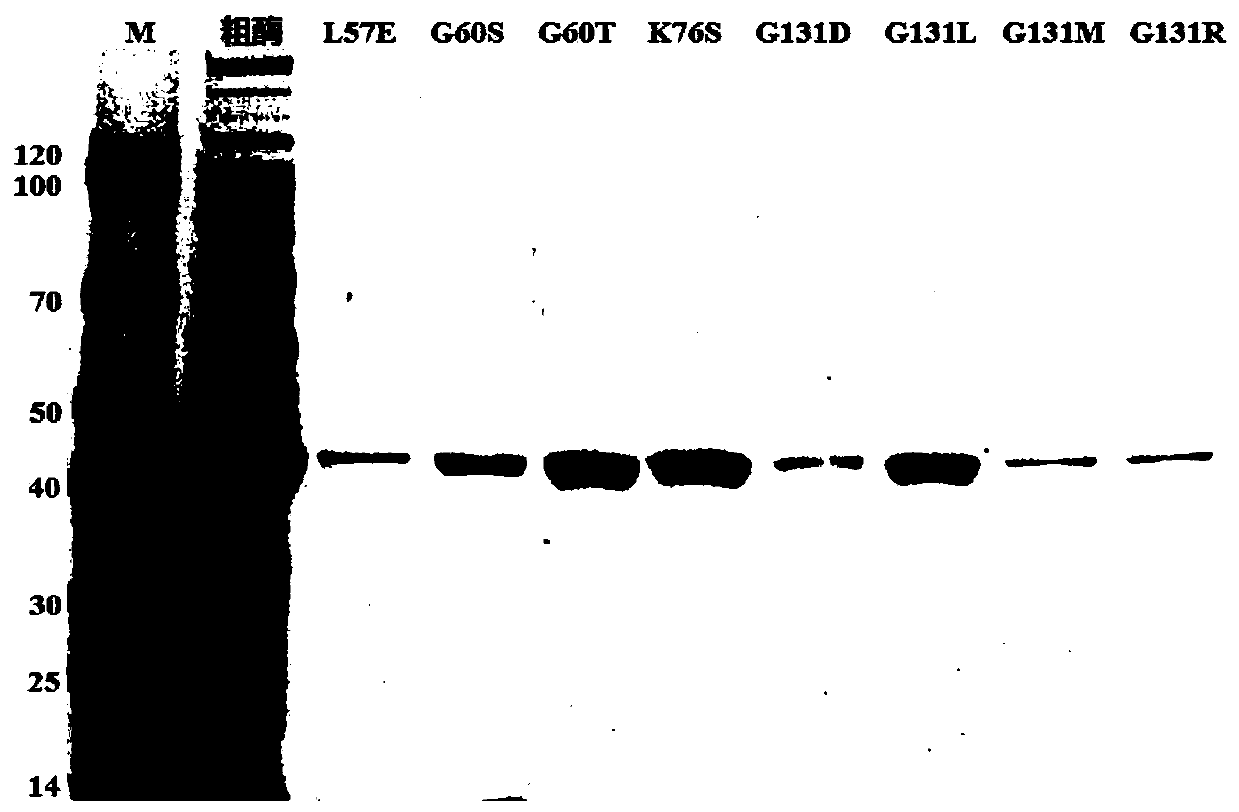
[0033] (2) Preparation of crude enzyme solution: construct a recombinant expression strain capable of expressing phenylalanine dehydrogenase with His-tag tag: the original sequence of phenylalanine dehydrogenase (PheDH) gene is from Bacillus nanh...
Embodiment 2
[0039] To construct a single point mutation (K76S) amino acid dehydrogenase mutant, to mutate the 76 amino acid, the specific steps are as follows:
[0040] (1) Introducing mutations: Design primers based on the nucleotides shown in SEQ ID NO. 02, and design forward and reverse primers containing different positions. The forward and reverse primers are as follows:
[0041] Upstream primer (SEQ ID NO.9): K76S-F CTTCGGTGGAGGTTCATCGGTCAT
[0042] Downstream primer (SEQ ID NO.10): K76S-R GAACCTCCACCGAAGTCAACATCT
[0043] After mixing the primers and the template plasmid, add high-fidelity Taq polymerase KOD-Plus to perform PCR amplification of the whole plasmid. After PCR, the PCR products are detected by electrophoresis, such as figure 1 Shown.
[0044] The experimental steps (2)-(3) are the same as the steps (2)-(3) of Example 1.
[0045] (4) Enzyme activity detection: The catalytic reaction system contains 100mM NADH, 0.4mol / L glycine-sodium hydroxide buffer solution (pH 10.5), the co-sol...
Embodiment 3
[0048] To construct a single-point mutation (M333D) amino acid dehydrogenase mutant, to mutate amino acid 333, the specific steps are as follows:
[0049] (1) Introducing mutations: Design primers based on the nucleotides shown in SEQ ID NO. 02, and design forward and reverse primers containing different positions. The forward and reverse primers are as follows:
[0050] Upstream primer (SEQ ID NO.17): M333D-FCAGATATCTACGGATGAAGCAGCA
[0051] Downstream primer (SEQ ID NO.18): M333D-R ATCCGTAGATATCTGGTTCTTTGT
[0052] After mixing the primers and the template plasmid, the high-fidelity Taq polymerase KOD-Plus is added to perform PCR amplification of the whole plasmid. After PCR, the PCR products are detected by electrophoresis.
[0053] The experimental steps (2)-(3) are the same as the steps (2)-(3) of Example 1.
[0054] (4) Enzyme activity detection: The catalytic reaction system contains 20mM NADH, 0.2mol / L glycine-sodium hydroxide buffer solution (pH 8.0), the co-solvent is 15% Tween...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com