In-vitro enzyme activity application of citrus-oxygen-methyltransferase CitOMT2
A technology of citrus oxymethylation and transferase, applied in the fields of genetic engineering technology and enzymology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] Example 1: Cloning of CitOMT2 full length
[0022] (1) Experimental method
[0023] 1. Extraction of RNA from the peel of 'Oukan' by CTAB method
[0024] Cut the fresh 'Oukan' peel into cubes, add liquid nitrogen to quick-freeze and grind; take 0.3g sample powder and add it to 4ml CTAB / β-mercaptoethanol extract preheated at 65°C in advance, vortex and mix well Continue heating at 65°C for 5 minutes; add 4ml of chloroform:isoamyl alcohol (24:1 / V:V) extract to the mixture, vortex and mix well, then centrifuge at 15°C and 10,000rpm for 10min; absorb the supernatant Liquid to a new centrifuge tube, add 4ml of the above extract, repeat the extraction, after centrifugation, draw the supernatant to a new centrifuge tube; add 1 / 4 volume of 10M lithium chloride solution to the obtained supernatant, Stand overnight at 4°C; the next day, centrifuge at 4°C, 10,000rpm for 30min, discard the supernatant; add 400μL of SSTE preheated at 65°C to the precipitate, tap to dissolve the pr...
Embodiment 2
[0033] Example 2: Expression and purification of CitOMT2 recombinant protein
[0034] (1) Experimental method
[0035] 1. Construction of target gene recombinant cells
[0036] According to the CitOMT2 full-length sequence, the primer pair SEQ:No.5 and SEQ:No.6 with the pET32a restriction site are designed, and the CitOMT2-T plasmid obtained in Example 1 is used as a template, according to the amplification shown in Example 1. The amplification system and program cloned the full-length sequence of CitOMT2 (without terminator), and constructed the CitOMT2-PET32a recombinant plasmid. The recombinant plasmid with correct sequencing was transformed into the E. coli expression strain BL21(DE3)pLysS(Promega), the positive recombinant cells were screened by ampicillin (Amp) resistance medium and colony PCR, and the corresponding bacterial solution was added to an equal volume of 50 % glycerol stored at -80°C.
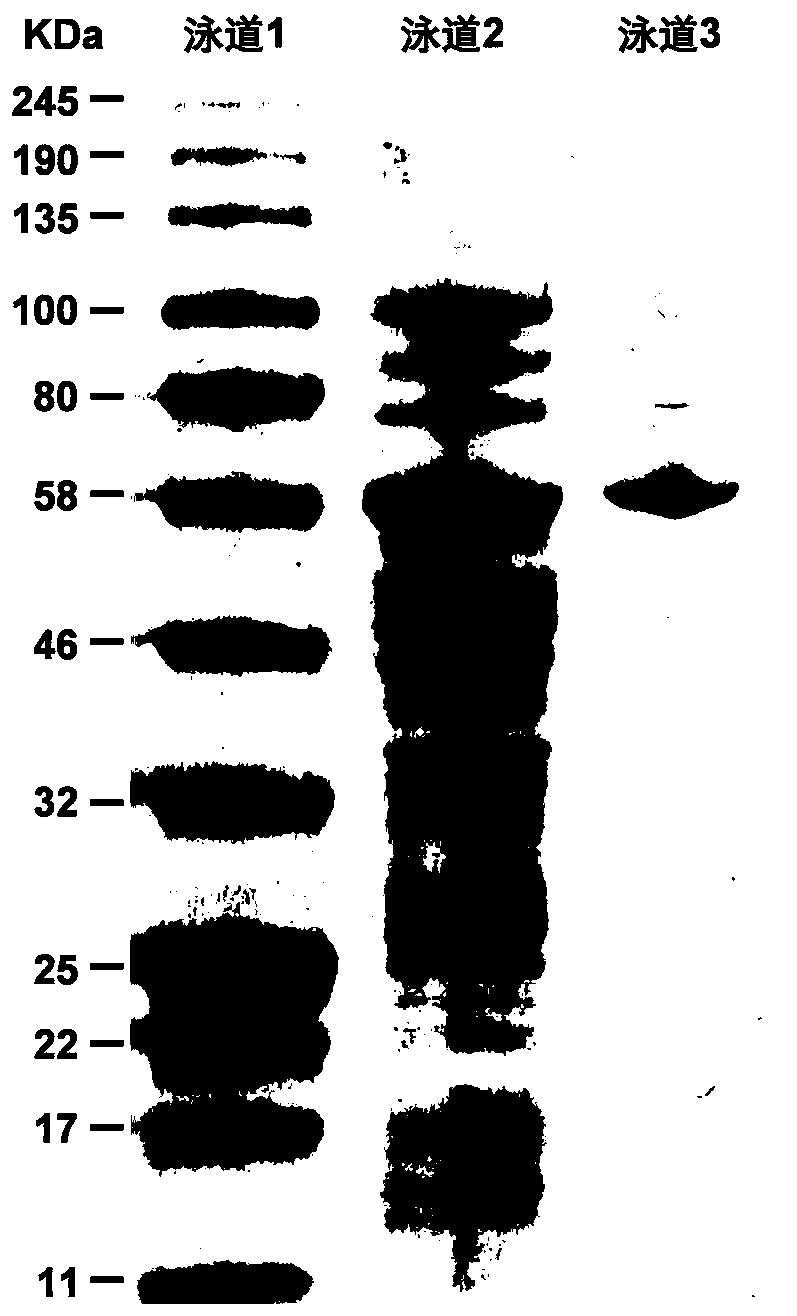
[0037] 2. Prokaryotic expression of target gene and purification of re...
Embodiment 3
[0041] Embodiment 3: CitOMT2 enzyme activity assay in vitro
[0042] (1) Experimental method
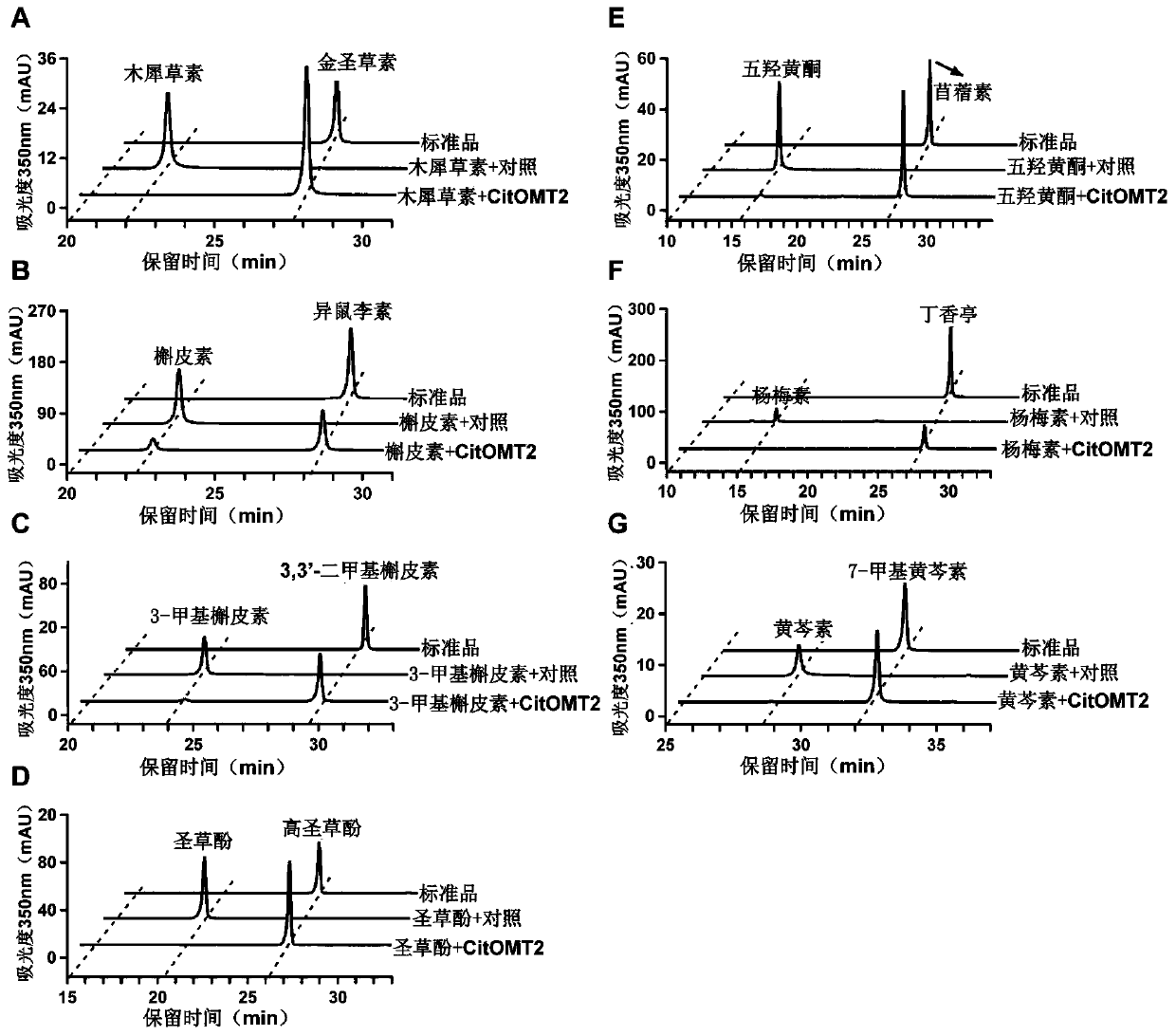
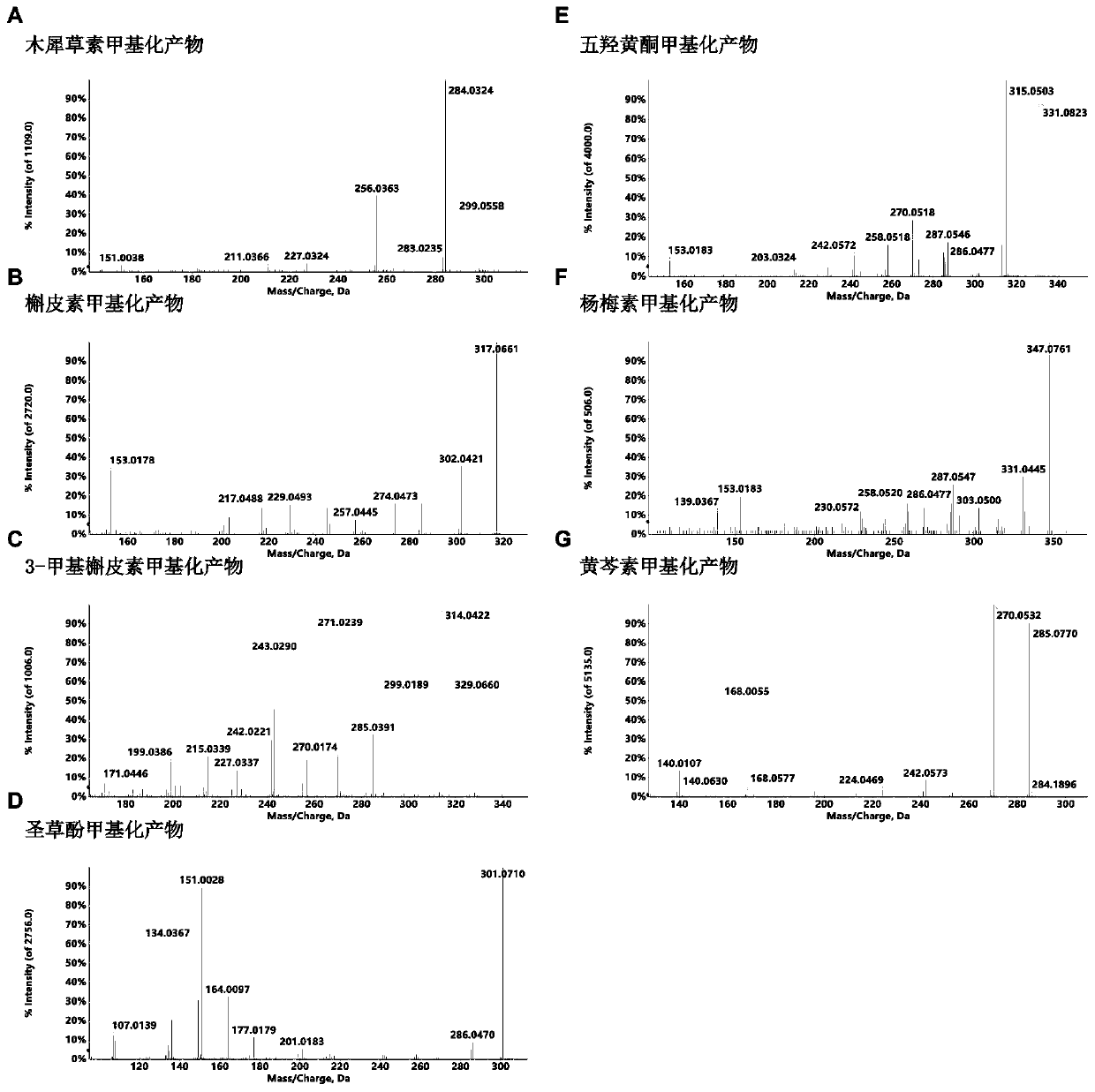
[0043] 1. CitOMT2 regioselectivity assay for different substrates
[0044] The regioselectivity of CitOMT2 recombinant protein was determined by seven substrates including luteolin, quercetin, 3-methylquercetin, eriodictyol, quercetin, myricetin and baicalein. The enzyme activity reaction system was 200 μL, including 25 μL of recombinant protein, 1 mM of S-adenosylmethionine, 200 μM of substrate, and the reaction buffer was Tris-HCl (pH 8.0). After the reaction solution was mixed, it was incubated in a water bath at 37°C for 2 hours, and an equal volume of methanol was added to mix well to terminate the reaction. The supernatant was centrifuged and passed through a 0.22 μm organic filter membrane for subsequent high-performance liquid chromatography (HPLC) and mass spectrometry analysis.
[0045] HPLC analysis condition: Agilent 1260HPLC system; Sunfire C18ODS chromatographic colum...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com