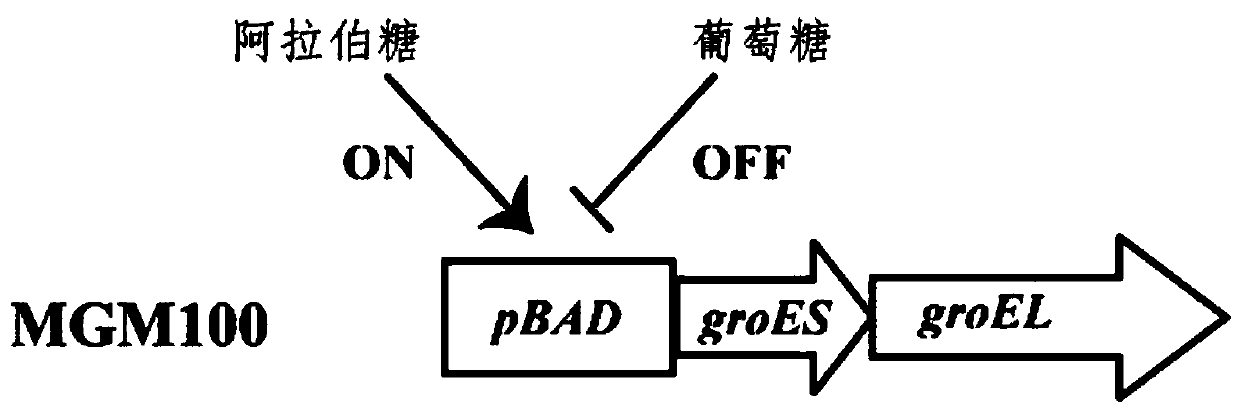
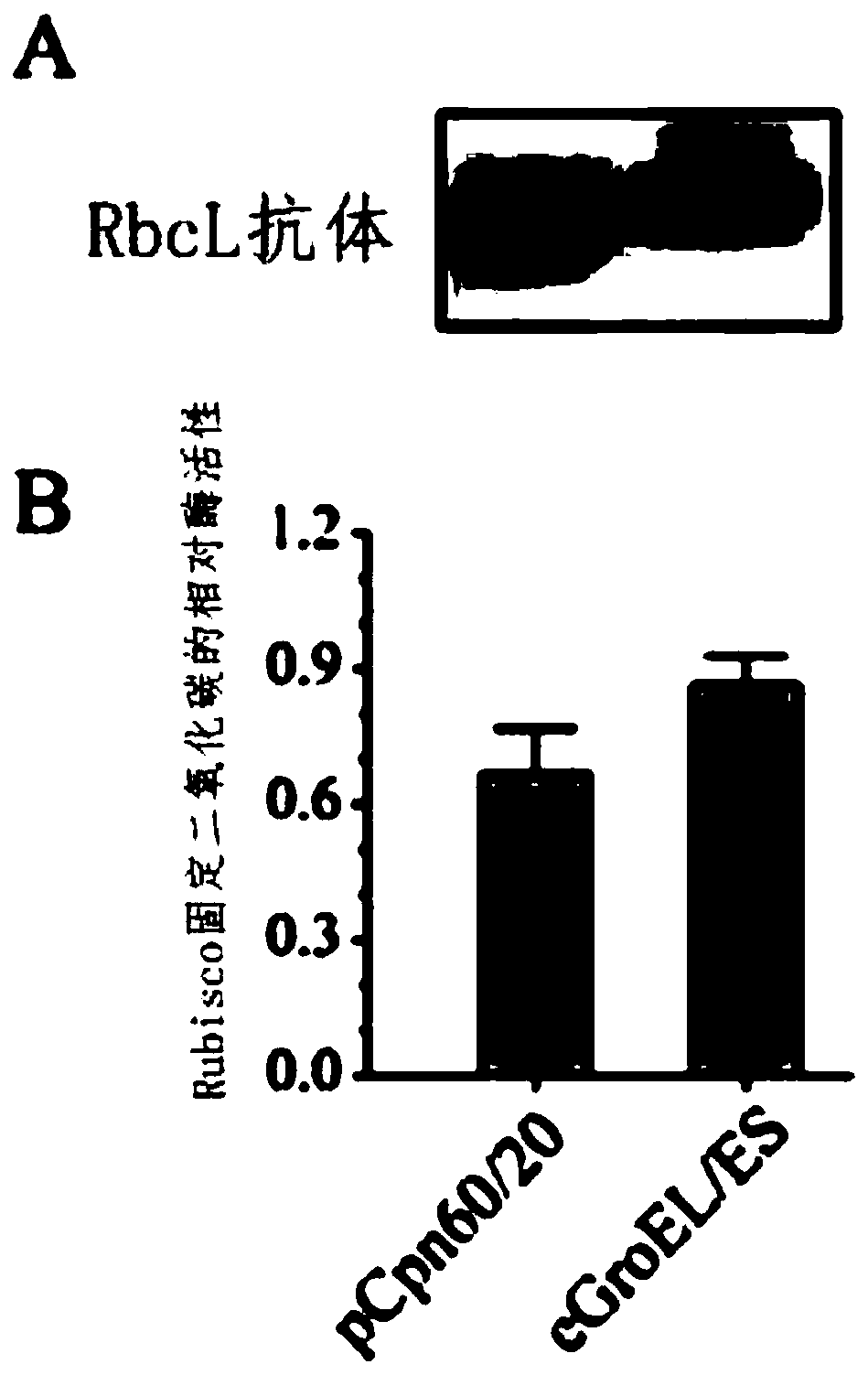
Application of Escherichia coli molecular chaperone GroEL/ES in synergic synthesis of plant Rubisco
A technology of molecular chaperone and Escherichia coli, which is applied in the biological field, can solve the problems of low efficiency and achieve the effect of avoiding dependence and simplifying the in vitro synthesis system
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Example 1 Plasmid 1: Construction of pTetlinker-AtRbcS-AtRbcL
[0041] 1. Construct the pTetlinker vector
[0042] 1. Construct the pTetlinker vector
[0043] 1. Use primers (pLCM1 For: cacggtcacactgcttccggta, pLCM1 Rev: gggatctcgaccgatgcccttgag) to PCR amplify plasmid pCDFDuet-1 (purchased from addgene: http: / / www.addgene.org / ) to obtain amplified fragment A (sequence 1 No. 1499- 3479), the amplified fragment A contains the expression cassette of the Escherichia coli CDF replicon (position 2574-3312 in sequence 1) and the spectinomycin resistance gene (position 1646-2434 in sequence 1).
[0044] 2. Use primers (pJKR-Tet For: agggcatcggtcgagatccc Gagtagggaactgccaggcatcaa, pJKR-Tet Rev3: gtgcttctcgcctgaggttCCGTGTGCTTCTCGCCTGAGGTT) to PCR amplify plasmid pJKR-H-TetR (purchased from addgene: http: / / www.addgene.org / ) to obtain amplified fragments B (sequence 2), the amplified fragment B comprises the expression cassette of the tetracycline inhibitory protein gene (sequenc...
Embodiment 2
[0068] Example 2 Plasmid 2: Construction of pT7linker1-Cpn60α1β1 / Cpn20
[0069] 1. pT7linker1 vector
[0070] 1. The artificially synthesized T7 promoter-lac operator-T7 terminator sequence shown in Sequence 5 (the sequence elements include T7 promoter (sequence 5 No. 1-19), lac operator (Sequence 5 No. 20-44 ) and T7 transcription terminator (sequence 5, 161-203)) to replace pQlinkN (Scheich, C., Kummel, D., Soumailakakis, D., Heinemann, U., Bussow, K., Vectors for co- expression of an unrestricted number of proteins, Nucleic Acids Research, 2007, Vol.35, No.6, e43.) in the Tac promoter-λt0 ter sequence (as shown in sequence 6) to obtain the pT7linker1 vector, the specific steps are as follows:
[0071] 1. Design primers (T7 box for: gtcttgaggggttttttgagtaacaacaccatttaaatgga, T7box rev: ctatagtgagtcgtattaacaattgaatctattataattgttatccgc) PCR amplification vector pQlinkN, obtain the amplified fragment H shown in sequence 7, and the amplified fragment H includes the LacI repress...
Embodiment 3
[0102] Example 3 Plasmid 3: Construction of pT7linker2-AtBSD2-AtRaf1-AtRaf2-AtRbcX2
[0103] 1. Construction of pT7linker2 vector
[0104] 1. Synthetic primers (chl-p15A for: agcggtatcagctcactcaaaggcacggtcacactgcttccg, chl-p15A rev: aatggtttcttagacgtcaggtggccacaggtgcggttgctggc) PCR amplification plasmid pACYC184 (purchased from addgene: http: / / www.addgene.org / ), The amplified fragment M shown in sequence 12 is obtained, and the amplified fragment M comprises the Escherichia coli p15A replicon (position 1215-2053 in sequence 12) and the chloramphenicol resistance gene (position 194-853 in sequence 12).
[0105] 2. Design primers (pT7linker1 for: cacctgacgtctaagaaaccatt, pT7linker1 rev: Cctttgagtgagctgataccgct) to amplify the pT7linker1 vector, and obtain the Escherichia coli Col1 replicon (sequence 7 No. 2672-3260) and the ampicillin resistance gene (Sequence 7 No. 3434-4291) amplified fragment N of the expression cassette.
[0106] 3. The amplified fragment M and the amplif...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com