Method, primers and kit for detecting PD-L1 gene expression quantity through real-time fluorescent quantitative PCR
A PD-L1 and gene technology, applied in the field of real-time fluorescence quantitative PCR detection of PD-L1 gene expression, primers and kits, can solve the problems of incomplete and accurate reflection of the expression of the gene to be tested, unstable expression, etc. Achieve the effect of eliminating instability, avoiding subjectivity, and improving sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0076] Example 1 Extraction of RNA / DNA from MDA-MB-231 cells, K562 cells and tumor tissue samples treated with interferon-γ
[0077] Using the AllPrep DNA / RNAMini Kit, the DNA / RNA extraction kit can extract up to 1*10 7 Cells or 30mg tissue.
[0078] Cells: cell number ≤1×10 7 , add 600μL Buffer RLTPlus; tissue: tissue ≤ 30mg, add 600μL Buffer RLTPlus. The lysate was then centrifuged at top speed for 3 minutes and the supernatant was carefully removed using a pipette.
[0079] Transfer to an Allprep DNA adsorption column (placed in a 2mL collection tube), and centrifuge at 10,000rpm for 30 seconds.
[0080] RNA extraction: add an equal volume of 70% ethanol to the centrifuged liquid from the above-mentioned Allprep DNA adsorption column, and mix well.
[0081] Pipette 700uL into the RNeasy adsorption column (placed in a 2mL collection tube), centrifuge at 10,000rpm for 15 seconds, and discard the waste liquid in the collection tube. Repeat until all RNA is collected.
[...
Embodiment 2
[0091] Embodiment 2 carries out reverse transcription reaction to extraction sample RNA / DNA
[0092] Configure the following mixture in RNA-free Microtube:
[0093] Table 1, cDNA reverse transcription system 1
[0094]
[0095] Place in a water bath at 65°C for 5 minutes, and cool on ice.
[0096] Prepare the following reaction solution in the above tube to a total volume of 20uL, and carry out the reverse transcription reaction.
[0097] Table 2, cDNA reverse transcription system 2
[0098]
[0099] Mix gently and place in a water bath at 45°C for 60 minutes.
[0100] Place in a water bath at 70°C for 15 minutes and cool on ice.
[0101] Obtain a cDNA / DNA solution sample for the next step of fluorescent quantitative PCR reaction.
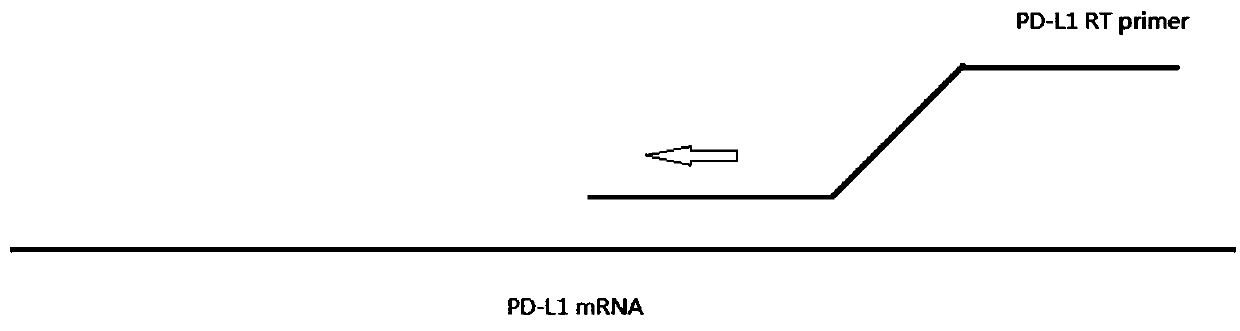
[0102] figure 1 is a schematic diagram of the reverse transcription reaction method, represented by figure 1 It can be seen that RNA is used as a template, PD-L1RT is used as a specific primer, and cDNA is obtained by reaction under th...
Embodiment 3
[0103] Example 3 Carry out dual-channel fluorescent quantitative PCR reaction and result calculation for cDNA / DNA samples
[0104] Configure the mixed solution according to the following reaction system (25uL / tube)
[0105] Table 3. Real-time quantitative PCR reaction system
[0106]
[0107] Fluorescent quantitative PCR reaction:
[0108] Table 4. Real-time quantitative PCR reaction conditions
[0109]
[0110]
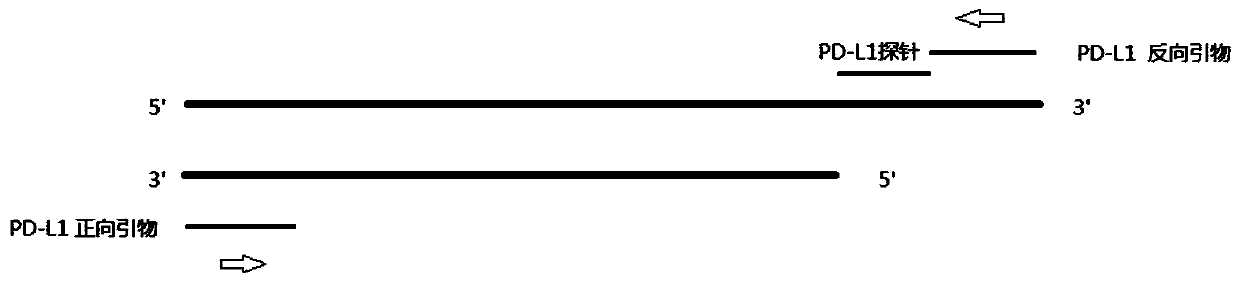
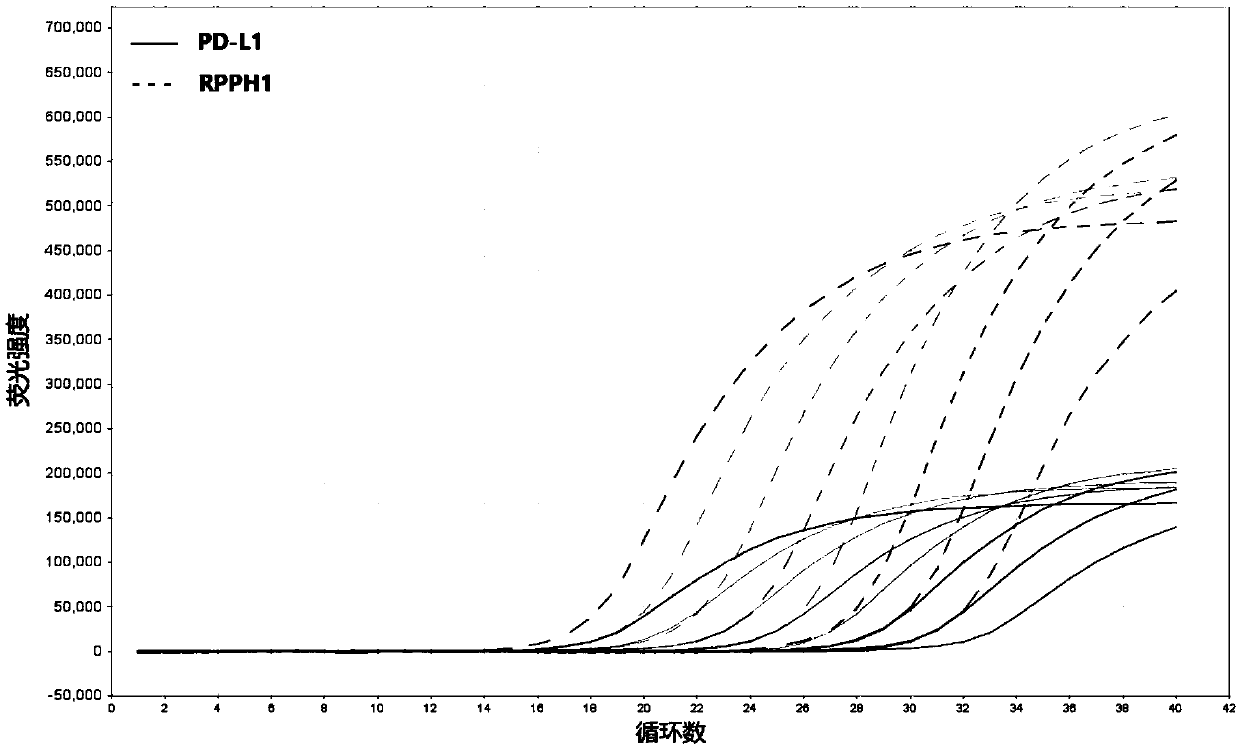
[0111] figure 2 It is the amplification method of PD-L1 cDNA fluorescent quantitative PCR, which is composed of figure 2 It can be seen that during PCR amplification, the 5'-3' exonuclease activity of Taq enzyme will digest and degrade the probe, so that the fluorescent emitting group and the fluorescent quenching group are separated, so that the fluorescent monitoring system can receive the fluorescent signal, that is Every time a DNA strand is amplified, a fluorescent molecule is formed, which realizes the complete synchronization of the accumulation...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com